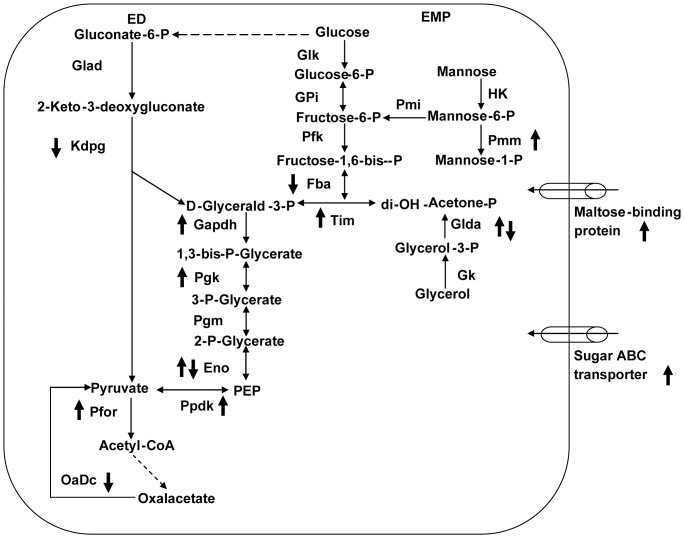
Figure 4. The possible network of central carbon metabolism in T. maritima.
The symbols, “↑, ↓, and ↓↑,” indicate the protein abundances up-, down-, or bell-shaped-regulation response to temperature change. Abbreviations: ED, Entner–Doudoroff pathway; EMP, Embden–Meyerhof–Parnas pathway; XynC, family 10 xylanase XynC; Glad, gluconate dehydratase; Kdpg, 2-dehydro-3-deoxyphosphogluconate aldolase; Glk, glucokinase; Gpi, glucose-6-phosphate isomerase; Pfk, phosphofructokinase; Fba, fructose-1,6-bisphosphate aldolase; HK, hexokinase; Pmi, phosphomannoisomerase; Pmm, phosphomannomutase; Tim, triosephosphate isomerase; Glda, glycerol dehydrogenase; Gk, glycerol kinase; Gapdh, D-glyceraldehyde-3-phosphate dehydrogenase; Pgk, 3-phosphoglycerate kinase; Pgm, phosphoglycerate mutase; Eno, enolase; Ppdk, pyruvate,orthophosphate dikinase; Pfor, pyruvate synthase subunit porA; OaDc, oxaloacetate decarboxylase; CoA, coenzyme A; PEP, phosphoenolpyruvate.