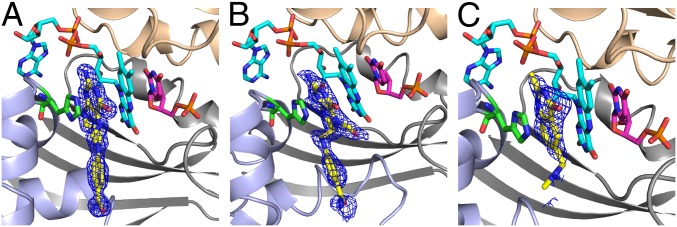
Fig. 3.
An active site view of crystal structures of TmFDTS in complex with FAD, dUMP, and folate derivatives. A view of the omit maps contoured at 3σ for CH2H4folate (A), folinic acid (B), and Raltitrexed (C). Ribbon drawings for the three protein chains constructing the active site (light gray, light blue, and wheat) and stick representation for FAD (cyan), dUMP (magenta), folate (yellow), and His53 (green). Note that the glutamate moiety of the tail is not defined in any of the crystal structures. The tail portion of the folate points to the solvent accessible region, and we also modeled the missing glutamic portion into the current structures and it fits well without any clashes with the protein residues. The PDB codes are 4GT9, 4GTA, and 4GTB for A, B, and C, respectively.