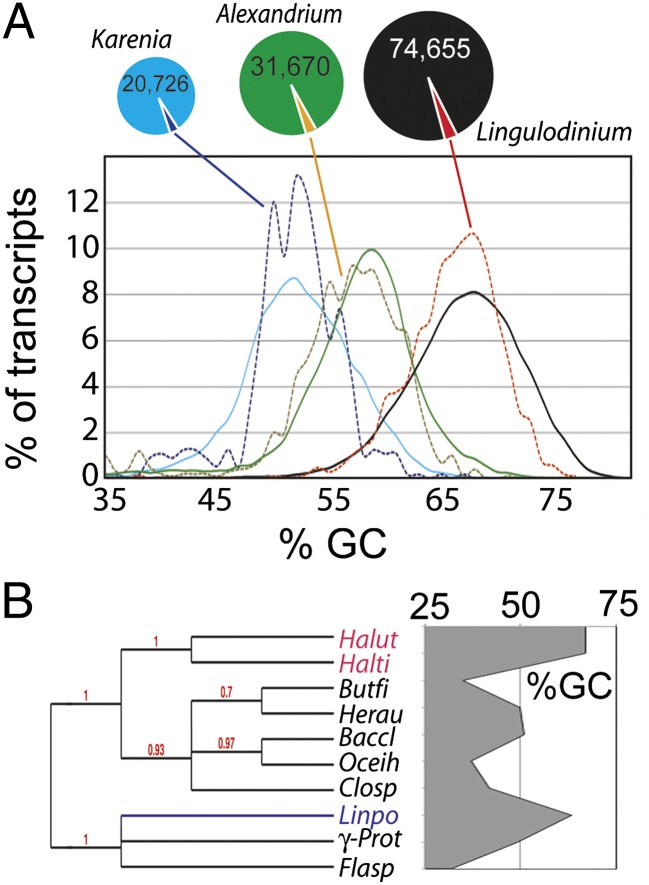
Fig. 3.
Bacteria-like sequences in the transcriptomes of different dinoflagellates have GC-contents commensurate with the host. (A) The GC-content of the unigene catalogs, as well as for potential bacterial sequences, was compared for datasets from Lingulodinium and two other dinoflagellate species, Alexandrium spp. and Karenia brevis. A similar proportion of potential bacterial sequences (3%) is found in all three datasets. Solid lines represent the entire dataset, dotted lines the dataset of potential bacterial sequences. (B) The GC-content of an arabinofuranosidase (JO761275) in L. polyedrum (Linpo) is higher than the more closely related eubacterial sequences (Butyrivibrio fibrisolvens, Herpetosiphon aurantiacus, Bacillus clausii, Oceanobacillus iheyensis, Clostridium sp., an unidentified γ-proteobacterium, and Flavobacteriales sp.) and is more similar to the more distantly related archeal sequences (Halorhabdus utahensis and Halorhabdus tiamatea).