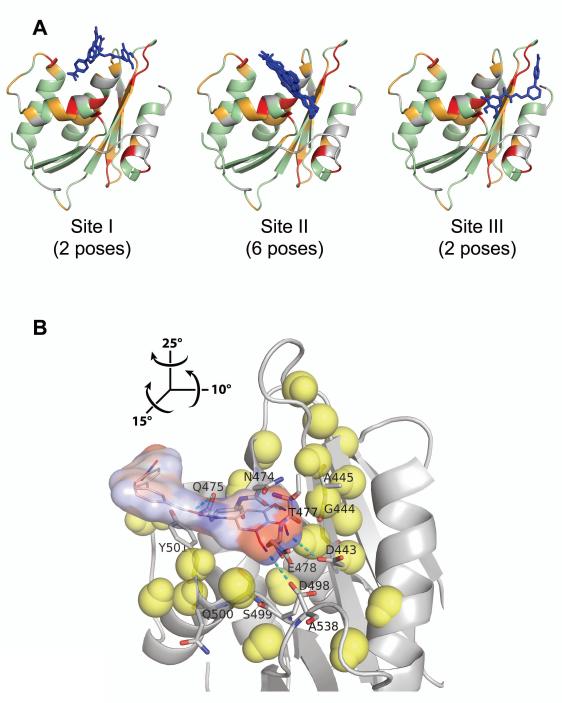
Figure 5.
Interaction of BHMP07 with RNH monomer mapped onto the crystallographic structure (PDB code, 3K2P, chain A). (A) Ten BHMP07 conformers with the highest docking scores at Site I, Site II, and Site III (see text). RNH residues (ribbon cartoon) that exhibit significant NMR chemical shift changes upon addition of 1.5× excess of BHMP07 (Figure 4A) are highlighted: ; ; ; undetected/unassigned residues are colored gray. The sampled BHMP07 poses (blue sticks, nonpolar hydrogens are omitted) were predicted via NMR-based molecular docking in the absence of divalent ions (see text). (B) Minimum energy conformer of BHMP07 docked at Site II on RNH (gray ribbon). The inhibitor pose is presented as the skeletal formula with superposed van der Waals contact surface, colored by partial atomic charges from −0.4 a.u. (red) to +0.4 a.u. (blue). Interacting RNH residues (Figure S7) are labeled and shown in stick representation with carbon, nitrogen and oxygen atoms denoted in gray, blue and red, respectively; predicted intermolecular hydrogen bonds are indicated (cyan dashes). Neighboring (< 11 Å) RNH backbone H and N atoms that exhibit significant chemical shift changes () are shown (yellow van der Waals spheres). Relative to panel (A), the view in (B) is rotated by 15°, −10° and −25° about the x, y and z-axes, respectively (see inset).