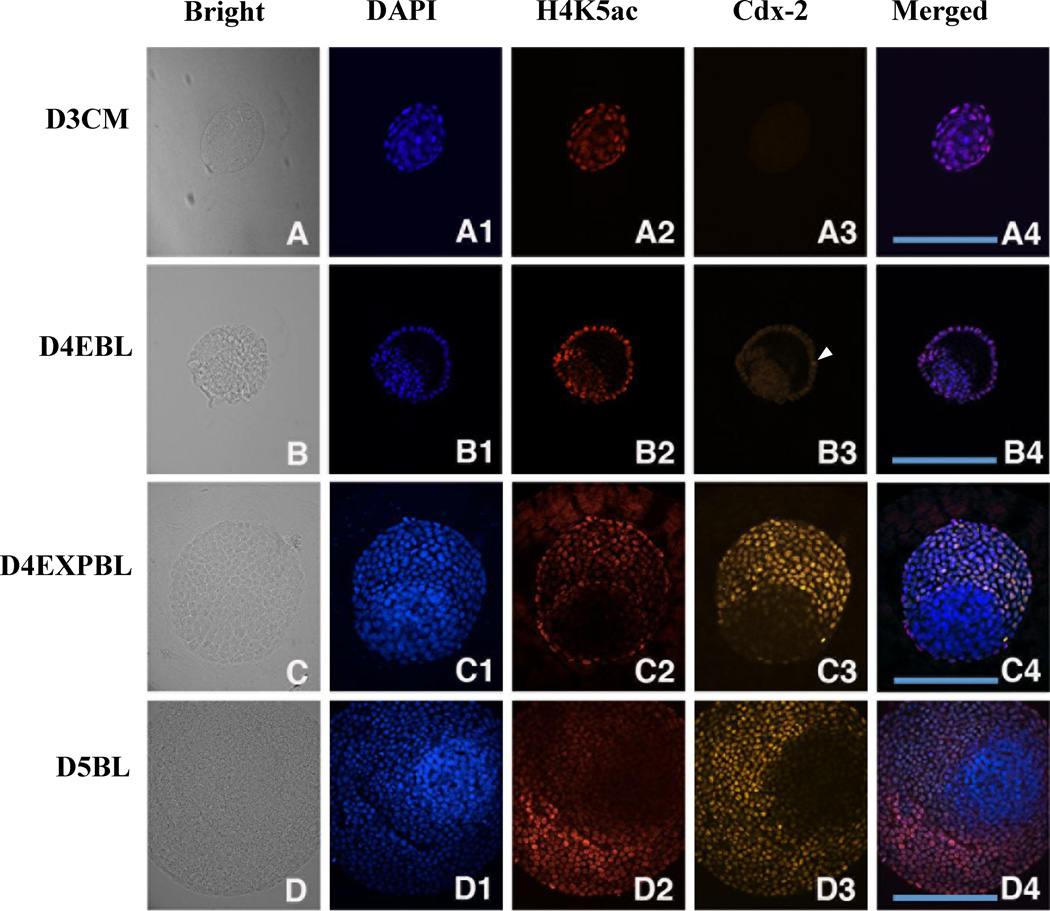
Figure 6.
Double staining of Cdx-2 and H4K5ac in in-vivo-derived rabbit embryos. A representative section across most of the area of ICM and TE was chosen for evaluation. The images of DNA (blue), H4K5ac (red) and Oct-4 (green) at different stages are shown. (A–A4) Cdx-2 signal was not detected in day-3 compact morulae, but hyperacetylated H4K5 was shown in the outside cells. (B–B4) Few Cdx-2 positive TE cells were initially found in day-4 early blastocysts (B3, arrowhead). Hyperacetylation of H4K5ac was continually shown in the TE lineage in contrast to hypo-acetylation in the ICM lineage. (C–C4) Cdx-2 signal exclusively appeared in the TE cells of day-4 expanded blastocysts accompanied with hyperacetylated H4K5 signal. (D–D4) A similar relationship between Cdx-2 and H4K5ac was found day-5 blastocysts. The expression patterns of Cdx-2 and H4K5ac across the TE and ICM regions were similar and overlapped in day-4 expanded blastocysts and day-5 blastocysts. D3CM = day-3 compact morulae; D4EBL = day-4 early blastocysts; D4EXPBL = day-4 expanded blastocysts; D5BL = day-5 blastocysts; D6BL = day-6 blastocysts; D7BL = day-7 blastocysts; ICM = inner cell mass; TE = trophectoderm. Bars = 200 µm.