Abstract
The ubiquitin proteasome pathway controls the cellular degradation of ~80–90% of the proteome in a highly regulated manner. In this pathway, E3 ligases are responsible for the conjugation of ubiquitin to protein substrates which can lead to their destruction by the 26S proteasome. Aberrant E3 ligases have been implicated in several diseases and are widely recognized as attractive targets for drug discovery. As researchers continue to characterize E3 ligases, additional associations with various disease states are being exposed. The availability of assays that allow rapid analysis of E3 ligase activity is paramount to both biochemical studies and drug discovery efforts aimed at E3 ligases. To address this need, we have developed a homogenous assay for monitoring ubiquitin chain formation using Tandem Ubiquitin Binding Entities (TUBEs). TUBEs bind selectively to polyubiquitin chains versus mono-ubiquitin thus enabling the detection of polyubiquitin chains in the presence of mono-ubiquitin. This assay reports on the proximity between the protein substrate and TUBEs as a result of polyubiquitin chain formation by an E3 ligase. This homogenous assay is a step forward in streamlining an approach for characterizing and quantitating E3 ligase activity in a rapid and cost effective manner.
Keywords: Ubiquitin, E3 ligase, Assay, MuRF1, TRIM25, Drug Discovery
Introduction
Ubiquitylation is a post translational modification that controls many aspects of cellular physiology. Ubiquitin (Ub) is a 76 amino acid polypeptide that is conjugated to target proteins and affects a protein’s cellular role in a myriad of ways. The original and most widely studied role for ubiquitin is its involvement in a protein’s stability; however it is also involved in regulating protein localization, oligomerization, activity, and function [1–3]. Ubiquitin modifications play a complex role in the cell, and new technologies are needed for dissecting and understanding these functions.
The conjugation of ubiquitin to a substrate protein involves a sequential three step enzymatic cascade that covalently attaches the C-terminal glycine residue of ubiquitin to an amine group, typically a lysine residue [4]. First, an E1 activating enzyme activates ubiquitin’s C-terminus in an ATP-dependent reaction. Next, the E1 interacts and transfers the activated ubiquitin to an E2 conjugating enzyme, where the E2 forms a thioester bond with ubiquitin. Lastly, an E3 ligase interacts with the E2-ubiquitin complex and a specific substrate protein, enabling the transfer of ubiquitin to the substrate. A substrate can be monoubiquitylated at a single lysine residue or multi-monoubiquitylated at several lysine residues. Additionally, it is possible to form a polyubiquitin chain by attaching additional ubiquitin molecules. There are seven lysine residues and the N-terminal amino group present in ubiquitin which can be used for chain assembly, each of which has a specific outcome.
Polyubiquitin chains on a target protein have been extensively characterized and many have been shown to be directed to the proteasome for degradation. The proteasome is a complex structure which hydrolyses 80–90% of soluble proteins into short oligopeptides and serves a critical role in cellular physiology [5]. The approval of bortezomib (VELCADE), a proteasome inhibitor, for the treatment of cancer, validated modulation of the ubiquitin pathway as an effective therapeutic approach [6]. However, toxicities associated with proteasome inhibition limit its clinical application. These toxicities are most likely a result of the fact that proteasome inhibition will impact the stability of thousands of proteins. In contrast, targeting enzymes upstream of the proteasome with small molecules should impact a smaller population of proteins and will display less toxicity than proteasome inhibition.
There are more than 600 predicted E3 ligases and each one is responsible for the ubiquitylation of a small number of proteins making these enzymes attractive targets for the treatment of numerous pathological conditions. The complexity of the ubiquitin pathway has made it challenging to develop technologies to readily study, characterize and quantitate E3 ligase activity.
Tandem Ubiquitin Binding Entities (TUBEs) are linear fusions of ubiquitin binding domains (UBAs) that bind polyubiquitin chains with dissociation constants in the nanomolar range. The nanomolar affinity of TUBEs for polyubiquitylated proteins allows for the detection of E3 ligase activity relative to free monomeric ubiquitin [7]. TUBEs are becoming established as one of the more versatile research tools for the exploration of the ubiquitin pathway.
Time Resolved Fluorescence Resonance Energy Transfer (TR-FRET) detection was selected as the optimal means to quantitate the binding of TUBEs to the polyubiquitin chains as the product of the E3 ligase reaction. FRET is a technique that relies on proximity dependent transfer of resonance energy from a donor fluorophore to an acceptor fluorophore. In FRET, a donor fluorophore is excited by incident light, and if an acceptor is in close proximity, the excited state energy from the donor can be transferred [8]. Energy that is emitted from a fluorophore is characteristically in the form of light at a specific wavelength. Because of this energy transfer, molecular interactions between biomolecules can be assessed by coupling each partner with a distinct fluorescence label and detecting the level of energy transfer. This method utilizes the validated TR-FRET lanthanide chemistry. The lanthanides have a long fluorescence lifetime and help avoid limitations associated with traditional FRET chemistries, where background fluorescence from sample components such as buffers, proteins, chemical compounds and cell lysate interfere with the fluorescent readout. Lanthanide donor/acceptor pairs have been optimized for qualitative and quantitative purposes, which includes spatial resolution, distance range, and sensitivity.
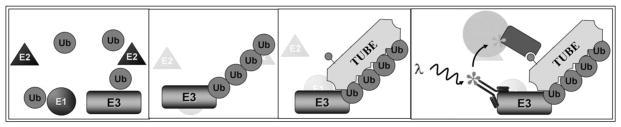
We applied TR-FRET and TUBEs to develop a novel, homogenous E3 ligase activity assay. Specifically, we monitored the proximity of the autoubiquitylated E3 ligase and our detection reagent, biotinylated TUBEs in an E3 dependent polyubiquitin chain formation assay (Figure 1). It has previously been shown that HTRF® technology (Cisbio) is capable of detecting E3 ligase activity for a few E3 ligases, such as Hdm2 and E6AP [9, 10]. However, a key limitation associated with this method in our view is the use of biotinylated ubiquitin. In an approach that intentionally avoids altering ubiquitin and deviating from its native structure, we use biotinylated TUBEs as our proximity detection reagent. In this manner, we can use native ubiquitin and TUBE detection to report on E3 dependent ubiquitin chain formation.
Figure 1.
Schematic model illustrating the homogenous E3 ligase assay platform.
Herein, we will provide an analysis of how to quantitate E3 ligase activity using TUBEs in a homogenous manner using CisBio HTRF® technology. We evaluated time and dose dependency, and the homogeneous nature of this assay allows rapid profiling of E3 ligase activity. MuRF1 and TRIM25 were analyzed in this manner. MuRF1 is an ubiquitin E3 ligase associated with muscle wasting, upregulated in numerous atrophy models [11, 12]. MuRF1 targets myofibrillar proteins for degradation and is itself a novel target to treat myopathy. TRIM25 also demonstrated ubiquitylation activity in this assay. TRIM25 is upregulated in response to estrogen, and it is thought to mediate estrogen actions in breast cancer as a primary response gene [13, 14]. This novel assay was employed to study the activity of these two E3 ligases.
Materials and Methods
Materials
Unless stated otherwise, all reagents were obtained from Sigma (St. Louis, MO) and were at a minimum of reagent grade or better. Plasmids encoding Ube1 and MuRF1 cDNAs were purchased from Open Biosystems; while other vectors and proteins were generous gifts (please see acknowledgements).
Cloning and Protein Expression
The cloning, expression, and purification from BL21 (DE3) bacteria of ubiquitin, 6his-Ube1, 6his-UbcH5c, GST-MuRF1, and GST-TRIM25 were performed using standard molecular biology techniques [15]. All constructs were fully sequenced to verify wildtype sequence.
E3 Ligase Autoubiquitylation
Nunc 384-well white polypropylene microtiter plates (Thermo Fisher Scientific) were used for autoubiquitylation assays. In a typical assay, varying concentrations of an E3 ligase were incubated with 5 nM Ube1, 100nM E2, 0.6 μM Ubiquitin, and 0.2mM ATP in a 10 μl reaction buffer (50mM Tris-HCl, pH8.0, 5mM MgCl2, and 1mM β-mercaptoethanol) in each well. After incubation at room temperature for a fixed period of time within the initial linear range (15–30 minutes for MuRF1), detection reagents were added. E3 autoubiquitylation was detected by the addition of 10nM biotinylated TUBE1 (Lifesensors Inc.) and HTRF® detection fluorophores anti-GST K (cryptate) & Streptavidin-XL665 (phycobilliprotein pigment) in detection buffer at a concentration recommended by the manufacturer (1:500). The detection buffer contained potassium fluoride (KF), where the final concentration was 800mM in 20ul. The KF concentration used was determined to give greatest signal to background (data not shown). Incubate for 1 hour at room temperature. The relative levels of ubiquitylated product were determined by an increase in fluorescence intensity using a fluorescence plate reader (Perkin Elmer Envision).
Kinetic E3 Ligase Autoubiquitylation
Nunc 384-well white polypropylene microtiter plates (Thermo Fisher Scientific) were used for autoubiquitylation assays. In a typical assay, varying concentrations of an E3 ligase were incubated with 5 nM Ube1, 100nM E2, 0.6 μM Ubiquitin, and 0.2mM ATP in 50mM Tris-HCl, pH8.0, 5mM MgCl2, and 1mM β-mercaptoethanol in each well with detection reagents. Detection reagents present were 1nM biotinylated TUBEs, 1nM Streptavidin-Tb (Terbium donor), and 40nM anti-GST D2 (acceptor) in a final volume of 20ul. Relative levels of ubiquitylated product were determined by a fluorescence plate reader (Perkin Elmer Envision).
Analyzing data in Excel
Using Microsoft Excel, CisBio’s ratiometric calculation was applied using raw data from the plate reader. HTRF® emission was measured at two wavelengths, Em1:595/60nm, Em2:665/7.5nm using an excitation wavelength of Ex: 340/60nm. The HTRF® ratio is calculated using the equation: Ratio= [(A665nm)/(B595nm)] × 104. The multiplying factor of 104 is used for the purpose of data handling. The ratio for each well within an assay is calculated and the mean and standard deviation of replicates determined using these values.
Results
Tandem Ubiquitin Binding domains recognize polyubiquitin chains
Previously, Ubiquitin Binding domains (UBAs) were employed to capture polyubiquitin chains when immobilized in a 96-well plate in a sandwich- based ELISA protocol [16]. Hjerpe et al. demonstrated that four linearly-fused UBAs appropriately named Tandem Ubiquitin Binding Entities (TUBEs) efficiently bind polyubiquitin chains with superior affinity when compared to a single ubiquitin binding domain [7]. To enhance throughput, we recognize that TUBEs need not be immobilized, and can bind polyubiquitin chains freely in solution [7]. TUBEs have outperformed single UBA domains, exhibiting a better signal to background ratio in our assays and for this reason our current assay focuses on the use of TUBEs in a homogenous E3 ligase assay (Data not Shown).
We utilized CisBio’s technology to monitor autoubiquitylation of GST-tagged MuRF1 and TRIM25. A schematic model in Figure 1 illustrates an E3 that undergoes autoubiquitylation in the presence of E1, E2 enzymes, ubiquitin, and ATP. Biotinylated TUBE1, (LifeSensors Inc.) added with a donor-labeled anti-GST antibody independently bind to the reaction product. TUBEs bind the ubiquitin chain when the GST-tagged E3 autoubiquitylates. Streptavidin-Acceptor (CisBio) is added and adheres to the biotinylated-TUBE1, bringing the acceptor fluorophore within close proximity of the donor-labeled antibody. This proximity allows for the generation of a signal, following the excitation of the donor fluorphore; the magnitude is proportional to the level of ubiquitylation.
TUBEs detect E3 ligase dependent ubiquitylation
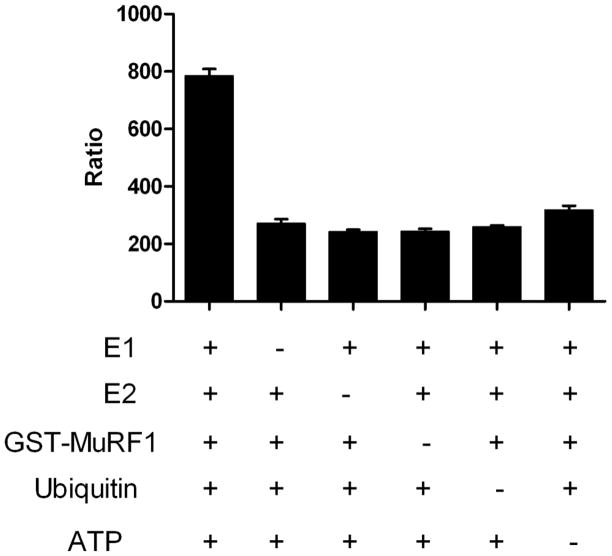
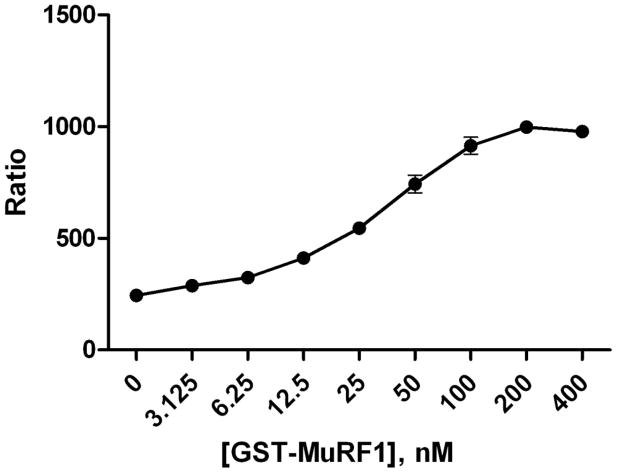
To test this assay, ubiquitin reactions consisted of a titration of GST-MuRF1 (3–400nM) with E1, E2, ATP and ubiquitin in a 10μl reaction. Detection reagents, TUBEs with anti-GST Europium and Streptavidin-XL665 were then added at 2x their final concentration. Following that addition, the plates were incubated at room temperature for an hour and then read on a PerkinElmer Envision plate reader with parameters recommended by CisBio. We used CisBio’s ratiometric measurement to analyze and process our TR-FRET data. As expected, MuRF1 autoubiquitylation was observed only when E1, E2, MuRF1, ubiquitin, and ATP were present (Figure 2). Figure 3 demonstrates that MuRF1 exhibited dose dependent polyubiquitylation. These data demonstrates that this assay strategy can be used to measure E3-dependent ubiquitylation of proteins where the signal to background is greater than 3.
Figure 2.
Assessment of MuRF1 autoubiquitylation activity. Ubiquitylation reactions conducted, where individual components (MuRF1, E1 (Ube1), E2 (UbcH5c), ubiquitin, and ATP) are absent as noted. Data are mean SD of triplicate determinations.
Figure 3.
Dose dependent E3 ligase activity. Ubiquitylation reactions with increasing concentration of the E3 ligase GST-MuRF1 correlates with an increased signal (Ratio) as detected in our homogenous assay platform. Data are mean SD of triplicate determinations.
Kinetic evaluation of ubiquitylation with TUBEs
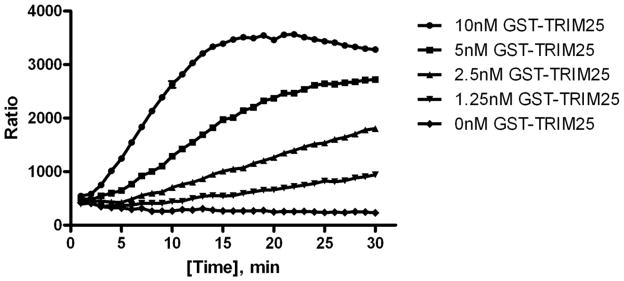
In Figure 4, we demonstrate time and dose dependency with the E3 ligase TRIM25, read in real time. In this assay, we switched the TR-FRET detection pair from Europium to Terbium (Tb) and XL665 to D2 after optimizing our protocol. In this format, FRET detection can be read in the absence of KF. Streptavidin was also labeled with the donor Tb instead of the acceptor. This minimized background interference since a low amount of streptavidin is needed for the assay, and therefore minimal amounts of Tb were used. The anti-GST antibody was then labeled with D2. This final arrangement led to achieving better signal strength over background (~8 fold) and laid the foundation for studying ubiquitylation kinetically.
Figure 4.
Dose and Time dependent E3 ligase activity. Varying concentrations of the E3 ligase GST-TRIM25 were incubated with ubiquitylation enzymes along with detectection reagents, 1nM TUBEs, 1nM streptavidin-Tb and 40nM anti-GST D2; and read kinetically.
In Figure 4, various concentrations of GST-TRIM25 were incubated with the ubiquitin reaction components along with the detection reagents, 1nM TUBEs, 1nM Streptavidin-Tb, and 40nM anti-GST D2. The assay exhibited dose-dependency at low nanomolar concentrations of E3 ligase with a linear increase in signal over time. This assay was conducted in the presence of detection reagents during the enzymatic reaction, allowing for a kinetic evaluation of E3 ligase activity in real-time. Advantages of the kinetic method over an end point method such as an ELISA, are shorter analysis time and reduced effects of interfering substances [17].
Discussion
Novel in vitro E3 assays are constantly being sought to improve the sensitivity and monitor activity under increasingly physiological conditions. To address this need, we developed a quantitative homogenous in vitro assay for ubiquitin E3 ligases, using Tandem Ubiquitin Binding Entities (TUBEs). Unlike previous methods we developed [16], this assay is a kinetic homogenous approach that utilizes unmodified ubiquitin. The advancement of this E3 assay format is that it monitors in real time and is highly adaptive to various E3 enzymes and potentially to substrates, allowing drug discovery to progress for known and emerging E3-mediated disease processes.
Previous experiments demonstrated that Tandem Ubiquitin Binding Entities (TUBEs) were able to bind polyubiquitin chains selectively with respect to free ubiquitin [7]. Subsequently, we were able to utilize this observation to develop an E3 ligase platform which reported dose and time dependent E3 autoubiquitylation in a homogenous manner. The utility of the assay platform was extended by the observation that it can also be used to kinetically determine E3 activity in real time. For these reasons, we conclude that this assay platform is an effective tool for characterizing autoubiquitylation activity of many ubiquitin E3 ligases.
Highlights of the paper include.
Demonstration of the assays’ capability to detect polyubiquitin chains relative to free ubiquitin using TUBEs by TR-FRET.
Profiling ubiquitin E3 ligases, demonstrating dose dependency in a homogenous manner.
Exhibiting kinetic analysis of E3 ligase activity in real time.
Given the broad interest in regulation of Ub/UBL mediated pathways and ubiquitin E3 ligases in particular, we believe that these methods and associated data will be of great interest to the readership of BBA.
Acknowledgments
We would sincerely like to thank Dr. Arthur Haas for his help and contributions. This work was supported by National Institute of Health (NIH) grant to Progenra Inc. (GM097827).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hershko A, Ciechanover A. The ubiquitin-proteasome pathway. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;16(2):119–26. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Ciechanover A. The ubiquitin-proteasomal pathway: on protein death and cell life. Embo J. 1998;17(24):7151–7160. doi: 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bross PF, Kane R, Farrell AT, Abraham S, Benson K, Brower ME, Bradley S, Gobburu JV, Goheer A, Lee SL, Leighton J, Liang CY, Lostritto RT, McGuinn WD, Morse DE, Rahman A, Rosario LA, Verbois SL, Williams G, Wang YC, Pazdur R. Approval summary for bortezomib for injection in the treatment of multiple myeloma. Clin Cancer Res. 2004;10(12 Pt 1):3954–64. doi: 10.1158/1078-0432.CCR-03-0781. [DOI] [PubMed] [Google Scholar]
- 7.Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10(11):1250–8. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selvin PR. The renaissance of fluorescence resonance energy transfer. Nat Struct Biol. 2000;7(9):730–4. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 9.Yabuki N, Watanabe S, Kudoh T, Nihira S, Miyamato C. Application of homogeneous time-resolved fluorescence (HTRFTM) to monitor poly-ubiquitination of wild-type p53. Comb Chem High Throughput Screen. 1999;2(5):279–87. [PubMed] [Google Scholar]
- 10.Kane SA, Fleener CA, Zhang YS, Davis LJ, Musselman AL, Huang PS. Development of a binding assay for p53/HDM2 by using homogeneous time-resolved fluorescence. Anal Biochem. 2000;278(1):29–38. doi: 10.1006/abio.1999.4413. [DOI] [PubMed] [Google Scholar]
- 11.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294(5547):1704–8. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 12.Clarke BA, Drujan D, Willis MS, Murphy LO, Corpina RA, Burova E, Rakhilin SV, Stitt TN, Patterson C, Latres E, Glass DJ. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 2007;6(5):376–85. doi: 10.1016/j.cmet.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Inoue S, Orimo A, Hosoi T, Kondo S, Toyoshima H, Kondo T, Ikegami A, Ouchi Y, Orimo H, Muramatsu M. Genomic binding-site cloning reveals an estrogen-responsive gene that encodes a RING finger protein. Proc Natl Acad Sci U S A. 1993;90(23):11117–21. doi: 10.1073/pnas.90.23.11117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horie K, Urano T, Ikeda K, Inoue S. Estrogen-responsive RING finger protein controls breast cancer growth. J Steroid Biochem Mol Biol. 2003;85(2–5):101–4. doi: 10.1016/s0960-0760(03)00209-7. [DOI] [PubMed] [Google Scholar]
- 15.Maniatis T, Fritsch EF, Sambrook J. Molecular cloning: A laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1983. [Google Scholar]
- 16.Marblestone JG, Suresh Kumar KG, Eddins MJ, Leach CA, Sterner DE, Mattern MR, Nicholson B. Novel approach for characterizing ubiquitin E3 ligase function. J Biomol Screen. 2010;15(10):1220–8. doi: 10.1177/1087057110380456. [DOI] [PubMed] [Google Scholar]
- 17.Srisawasdi P, Kroll MH, Lolekha PH. Advantages and disadvantages of serum cholesterol determination by the kinetic vs the end point method. Am J Clin Pathol. 2007;127(6):906–18. doi: 10.1309/1ENNJCJRN682F1HP. [DOI] [PubMed] [Google Scholar]