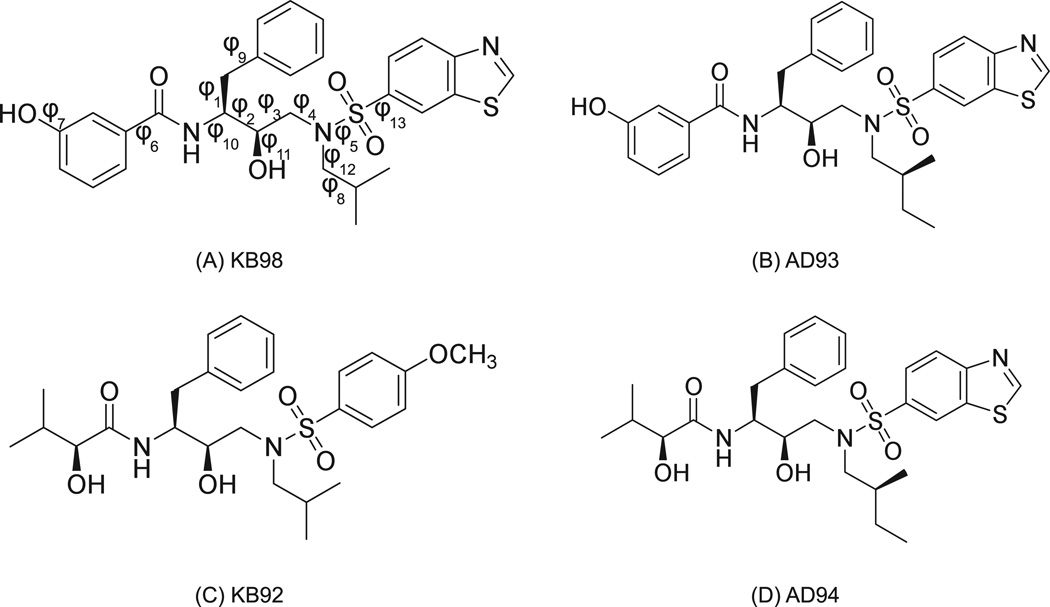
Figure 5. Chemical structures of HIV-1 protease inhibitors.
The four molecules shown were previously designed as candidate HIV-1 protease inhibitors.35 For the current work, idealized rotameric systems in which the exact energies of 50,000 rotameric states were generated in both bound and unbound states, as described in Methods. All torsional degrees of freedom for each inhibitor were rotamerized, and all other DOF (bonds, angles, impropers) were fixed to idealized values. In the bound state overall translations and rotations (external DOF) were also enumerated. Torsions about the bonds labeled in (A) correspond to numbering used in Figure 8.