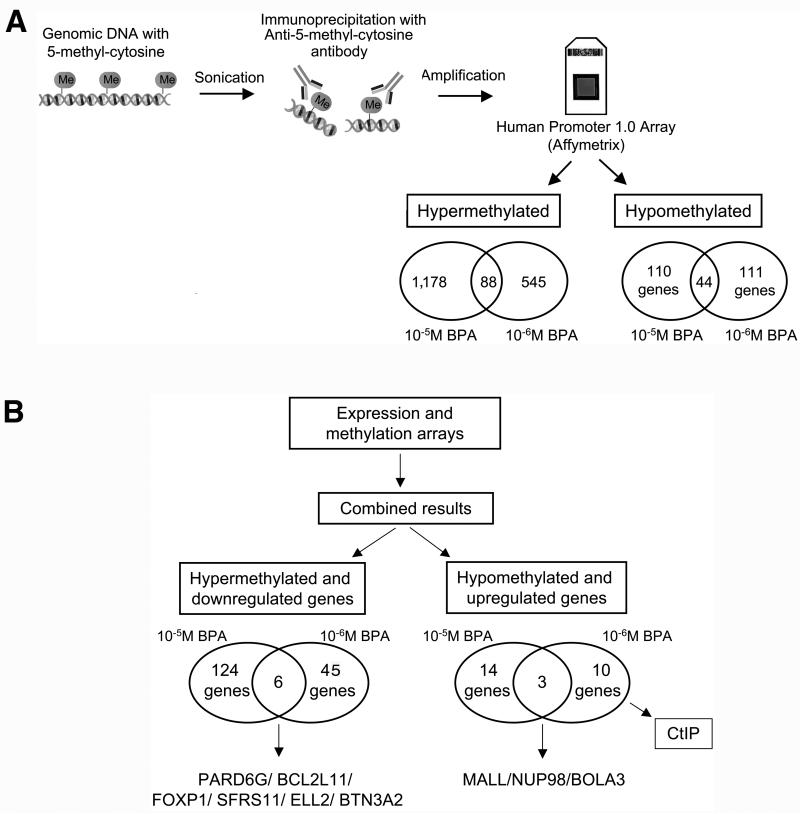
Figure 2. DNA methylation studies in MCF-10F cells exposed to BPA.
A) DNA was isolated from the cells and fragmented by sonication. Methylated DNA was isolated using an antibody against 5-methylcytidine and amplified, followed by hybridization using a promoter microarray to identify regions with altered methylation in the promoters. The Human Promoter 1.0R Arrays were used and hyper- and hypomethylated promoters were identified. Hypermethylated targets were sequences with significantly increased signals in the cells exposed to BPA relative to the control without treatment immunoprecipitated using the antibody against 5-methylcytidine; B) Combined results from the expression and DNA methylation arrays: The list of genes that were found hypermethylated by MeDIP-on Chip and the list of genes found down-regulated by expression arrays were compared. At the left, the number of genes hypermethylated and down-regulated is indicated for the cells exposed to 10-5M and 10-6M BPA. The same was done for genes hypomethylated and up-regulated; the number of genes hypomethylated and up-regulated is indicated for the cells treated with 10-5M and 10-6M BPA at the right.