Abstract
Inactivation of one X chromosome in female mammals (XX) compensates for the reduced dosage of X-linked gene expression in males (XY). However, the inner cell mass (ICM) of mouse preimplantation blastocysts and their in vitro counterparts, pluripotent embryonic stem cells (ESCs), initially maintain two active X chromosomes (XaXa). Random X chromosome inactivation (XCI) takes place in the ICM lineage after implantation or upon differentiation of ESCs, resulting in mosaic tissues composed of two cell types carrying either maternal or paternal active X chromosomes. While the status of XCI in human embryos and ICMs remains unknown, majority of human female ESCs show non-random XCI. We demonstrate here that rhesus monkey ESCs also display monoallelic expression and methylation of X-linked genes in agreement with non-random XCI. However, XIST and other X-linked genes were expressed from both chromosomes in isolated female monkey ICMs indicating that ex vivo pluripotent cells retain XaXa. Intriguingly, the trophectoderm (TE) in preimplantation monkey blastocysts also expressed X-linked genes from both alleles suggesting that, unlike the mouse, primate TE lineage does not support imprinted paternal XCI. Our results provide insights into the species-specific nature of XCI in the primate system and reveal fundamental epigenetic differences between in vitro and ex vivo primate pluripotent cells.
Keywords: X-inactivation, Embryonic Stem Cells, Blastocyst, Inner Cell Mass, Primates
Introduction
X chromosome inactivation is believed to be an essential mechanism regulating the dosage compensation of X-linked genes in eutherian mammals so that females with two X chromosomes do not overexpress X-linked genes compared to males (Lyon, 1961). XCI is initiated during early mouse preimplantation embryo development, where the paternally inherited X chromosome is silenced in early cleaving embryos. However at the blastocyst stage, paternal X is transiently reactivated in the ICM, resulting in two active X chromosomes (XaXa). However, paternally imprinted XCI is maintained in the mouse TE lineage (Hajkova and Surani, 2004; Okamoto et al., 2004). Random XCI takes place in the ICM lineage after implantation, at about the time of gastrulation, through epigenetic silencing involving XIST RNA coating of the inactive X in cis (Panning et al., 1997; Penny et al., 1996). Thus, somatic tissues in females are mosaic composed of two cell types expressing from one or the other X chromosome.
In contrast to this strict X gene dosage compensation mechanism in the mouse, approximately 15% of X-linked genes in humans escape XCI and are expressed biallelically in females (Carrel and Willard, 2005). Why and how these escape genes are transcribed from a largely inactivated X chromosome is not fully understood. In addition, the existence of paternally imprinted XCI in the TE lineage in humans remains controversial, where few studies reported conflicting findings (Moreira de Mello et al., 2010; Zeng and Yankowitz, 2003).
ESCs are in vitro pluripotent cell lines derived from the ICM of preimplantation blastocysts in several species, including mice, nonhuman primates, and humans (Evans and Kaufman, 1981; Martin, 1981; Thomson et al., 1998; Thomson et al., 1995). ESCs can be maintained and propagated indefinitely in a pluripotent state providing an unlimited supply of undifferentiated cells for cell replacement therapy. However, isolation of stable mouse female ESCs remains problematic due to frequent loss of one of the two X chromosomes (Zvetkova et al., 2005). In a few existing stable mouse XX ESCs, both X chromosomes remain active and XCI is initiated upon differentiation (Nichols and Smith, 2009).
In contrast to the mouse, isolation of male and female primate ESCs is equally efficient and loss of one of the two X chromosomes is relatively rare in human female ESCs. However, a majority of human female ESC lines appear to have undergone XCI in an undifferentiated state (Shen et al., 2008; Silva et al., 2008). Moreover, these human ESCs often exhibit monoallelic expression of X-linked genes, suggesting either imprinted XCI, as seen in the mouse TE lineage (Shen et al., 2008), or random XCI followed by the clonal selection of the one or another populations during ESC isolation and culture.
It remains unclear whether such fundamental differences between mouse and primate ESCs reflect species-specific differences in the tissue of origin. For example, XCI in human ESCs could simply reflect the pre-existing status in the parental ICMs. Alternatively, XCI may indicate epigenetic instability during isolation and long-term culture of human ESCs. Our recent study demonstrated that monkey ESCs are unable to contribute to chimeras upon injection into host blastocysts (Tachibana et al., 2012). However, transplanted ICMs formed viable fetuses while sharing the TE compartment with host blastocysts. These results necessitate further investigations into genetic and epigenetic mechanisms responsible for such drastic differences in developmental potential of primate ICMs vs. ESCs. Currently, few studies are available on X inactivation status and timing in human embryos (Okamoto et al., 2011; van den Berg et al., 2009). This is in large part, due to restrictions on human embryo research and the lack of relevant genetic markers that would allow discrimination of two X chromosomes.
To address this gap in the knowledge, we carried out a comprehensive analysis of XCI on a clinically relevant nonhuman primate model. We investigated allele specific expression and methylation of several X-linked genes in female rhesus macaque (Macaca Mulatta) blastocysts, focusing particularly on the ICM and TE. We also extended our studies to rhesus monkey ESCs derived from fertilized embryos or experimental pluripotent stem cells derived by reprogramming of somatic cells using somatic cell nuclear transfer (SCNT) or iPS (induced pluripotent stem) cell approaches.
Materials and methods
Animals
All animal procedures were approved by the Institutional Animal Care and Use Committee (AICUC) at the ONPRC/OHSU.
Production of monkey embryos, ICM and TE isolation and gender determination
Rhesus macaque embryos were generated by intracytoplasmic sperm injection (ICSI) and cultured to the blastocyst stage as described previously (Wolf et al., 2004). In vivo developmental competence of these embryos was previously demonstrated by birth of healthy rhesus offspring (Tachibana et al., 2012; Tachibana et al., 2009; Wolf et al., 2004). Intact ICMs from various stage blastocysts were isolated by immunosurgery (Mitalipov et al., 2006). In brief, zona pellucidae were digested by short (10 sec) treatment with 0.5% protease. Blastocysts were then incubated in anti monkey whole serum (Sigma) for 30 min at 37 °C, washed three times with culture medium and transferred into guinea pig complement (Sigma) for 30 min. Blastocysts were then gently pipetted with a small-bore pipette to disperse lysed TE cells and isolate intact ICMs. For the TE isolation, a zona-free blastocyst was held with a micropipette near the ICM. Next, a sequential laser pulse (Staccato laser www.hamiltonthorne.com) was fired across the boundary between the ICM and TE while the TE part was pulled away with a second pipette until complete separation. Unutilized ICM or TE cells were used for gender determination using PCR approach. Cells were collected into 0.2ml PCR tubes containing 4ul of PicoPure® DNA (Arcturus Bioscience) extraction buffer, and X- and Y-linked zinc finger protein genes (ZFX and ZFY) were amplified as previously described (Mitalipov et al., 2007). Female samples produced 1149 bp fragment while male samples contained an additional 771 bp fragment.
Derivation, culture and characterization of monkey iPS cells
Primary cultures of fibroblasts were established from rhesus macaque skin biopsies. Fibroblasts in the log growth phase were transduced with retroviral vectors carrying 4 transcription factors as previously described (Takahashi et al., 2007; Wu et al., 2009). Briefly, plasmids (pMXs-hocT4, pMXs-hSOX2, pMXs-hKLF4 and pMXs-hC-MYC, Cell Biolabs, Inc. San Diego, CA) were packaged into retroviral particles by transfection into Platinum-A Retroviral Packaging Cells using Fugene® HD Transfection Reagent (Roche, Indianapolis, IN). Transduction of fibroblasts was performed three times at 24 hr intervals, followed by seeding of cells onto feeder layers of mitotically inactivated mouse embryonic fibroblasts (mEFs) in ESC culture medium consisting of DMEM/F12 medium with high glucose, without sodium pyruvate and supplemented with 1% nonessential amino acids, 2 mM I-glutamine, 0.1 mM β-mercaptoethanol and 15% FBS (Mitalipov et al., 2006). The transduced cells were maintained at 37°C in 3% CO2, 5% O2 and balance N2 for up to 4 weeks or until colonies of cells with a morphology similar to ESCs appeared. Each colony was then individually isolated and manually propagated using standard ESC culture techniques as previously described (Byrne et al., 2007; Mitalipov et al., 2006; Sparman et al., 2009).
Expression of ESC markers in iPS cells was detected by immunocytochemistry as previously described (Mitalipov et al., 2006; Sparman et al., 2009). Primary antibodies for OCT4, SSEA-4, TRA-1-60 and TRA-1-81 were from Santa Cruz Biotechnology Inc. and NANOG was from R&D Systems.
Comparative microarray analysis of mRNA profiles in iPS cells and their IVF or SCNT controls was carried out using the Affymetrix Rhesus Macaque Genome array as previously described (Sritanaudomchai et al., 2010). RNA samples were converted to labeled cRNA and hybridized to GeneChip Rhesus Macaque Genome Arrays (Affymetrix, Inc.). Gene-Chip operating system version 1.4 software (Affymetrix) was used to process images and generated probe level measurements. Microarray data, including CEL and CHP files, can be accessed at the Gene Expression Omnibus (GEO: GSE36252), http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=zxcvbegqooeoalu&acc=GSE36252. Processed image files were normalized across arrays using the robust multichip average algorithm (Irizarry et al., 2003) and log transformed (base 2) to perform direct comparisons of probe set values between samples. GeneSifter (VizX Labs, Seattle, WA) microarray expression analysis software was used to identify differentially expressed transcripts. For a given comparison, IVF-derived ESCs were selected as the baseline reference, and transcripts that exhibited various fold change relative to the baseline were considered differentially expressed. To facilitate in-depth comparisons, processed image files were normalized with the robust multichip average algorithm and log transformed (base 2) using the StatView program. Corresponding microarray expression data were analysed by pairwise differences determined with the Student-t-test (P < 0.05).
Qualitative and Quantitative Reverse Transcription (RT)-PCR analysis
Total RNA was extracted from ESCs and preimplantation embryos using TRIzol® Reagent (Invitrogen) and PicoPure™ RNA extraction kit (Arcturus Bioscience), respectively. DNAse treated RNA was converted to cDNA using the SuperScript III first strand synthesis system for RT-PCR (Invitrogen) according to the manufacturer’s instructions. The first strand cDNA was amplified by PCR using XIST specific primers for RT or qPCR, as previously described (Sparman et al., 2009). The primers for G6PD qPCR were; F-5’ atgaaaggtgggatggagtg 3’ and R-5’ actgctggtggaagatgtca 3’. qPCR was performed using 7500 Fast system with Fast SYBR® Green PCR master Mix (Applied Biosystems). Expression of housekeeping GAPDH was used as a control.
Generation of heterozygous embryos and allele-specific expression analysis of XIST and X-linked genes
To generate heterozygote embryos, rhesus macaque males and females were pre-screened for SNPs within transcribed regions of XIST and X-linked genes as previously described (Fujimoto et al., 2006). Information for primers and SNP positions is available in Supplementary Material, Tables S1 and S2. Oocytes and sperm from animals carrying informative alleles were then collected and embryos were generated in vitro by ICSI as described previously (Wolf et al., 2004). RT-PCR amplicons were subcloned and expressed alleles were determined by direct sequence analysis of a minimum 10 individual clones. In brief, PCR reaction was carried out using PCR super mix high-fidelity DNA polymerase (Invitrogen) containing 0.5 µM of each primer (final volume 25 µl). PCR products were sequenced and the expressing alleles were determined using Sequencher v. 4.7 software (GeneCodes).
Bisulfite genomic sequencing
Genomic (g)DNA was extracted from ESCs and embryonic cells using Gentra PUREGENE® DNA Purification (Qiagen) or PicoPure® DNA kit (Arcturus Bioscience), respectively. gDNA was subjected to bisulfite treatment using a MethylCode™ Bisulfite Conversion Kit (Invitrogen, MECOV-50) according to the manufacturer’s instructions. In brief, 500ng of total gDNA was used for each reaction. For embryos, gDNA was pooled from up to twenty ICMs while for TEs three samples were combined to generate sufficient DNA for a single bisulfite reaction. Bisulfite sequencing PCR primers were designed with Methyl Primer Express Software v1.0 (Applied Biosystems). Information for primers and PCR conditions is available in Supplementary Material, Table S4. PCR reactions were performed using PCR super mix high-fidelity DNA polymerase (Invitrogen) containing 0.5 µM of each primer (final volume 25 µl). PCR products were then sub-cloned using TOPO® TA cloning kit. Fifteen to twenty-five colonies were randomly selected and sequenced. Sequences were then analysed using a web-based QUantification tool for Methylation Analysis (QUMA) (http://quma.cdb.riken.jp/). Methylation profiles were presented as a CpG map with white and black dots indicating unmethylated and methylated CpG dinucleotides, respectively.
Statistical Analysis
Statistical analyses were performed using ANOVA and Fisher’s PLSD with Statview Software (SAS Institute, Inc.) with statistical significance set at 0.05.
Results
XCI in Monkey Somatic Cells
To establish reliable markers of XCI, we initially analysed XCI status in somatic tissues derived from fetal and adult rhesus macaques. Based on the evidence that expression of XIST is critical for initiation of X chromosome silencing and early XCI readout (Diaz-Perez et al., 2005; Lucchesi et al., 2005), we initially detected transcripts of XIST. As expected, conventional reverse-transcription (RT)-PCR detected XIST expression in female somatic cells but not in males (Fig. 1A). We next screened and identified informative heterozygous fetal and adult tissues based on single nucleotide polymorphisms (SNPs) within transcribed regions for XIST. Information on SNP positions and primers is presented in supplemental information (Tables S1 and S2). Allele specific expression analysis demonstrated the presence of XIST transcripts from both X-chromosomes in female tissues (Table S3). This was an expected outcome assuming that female tissues represent a mixture of cells with random XCI. We also analysed allele-specific expression of X-linked ZNF41 and FMR1, genes shown to undergo silencing in humans (Carrel and Willard, 2005). Similar to XIST, transcripts from both X-chromosomes were detected in female tissues for both genes (Table S3).
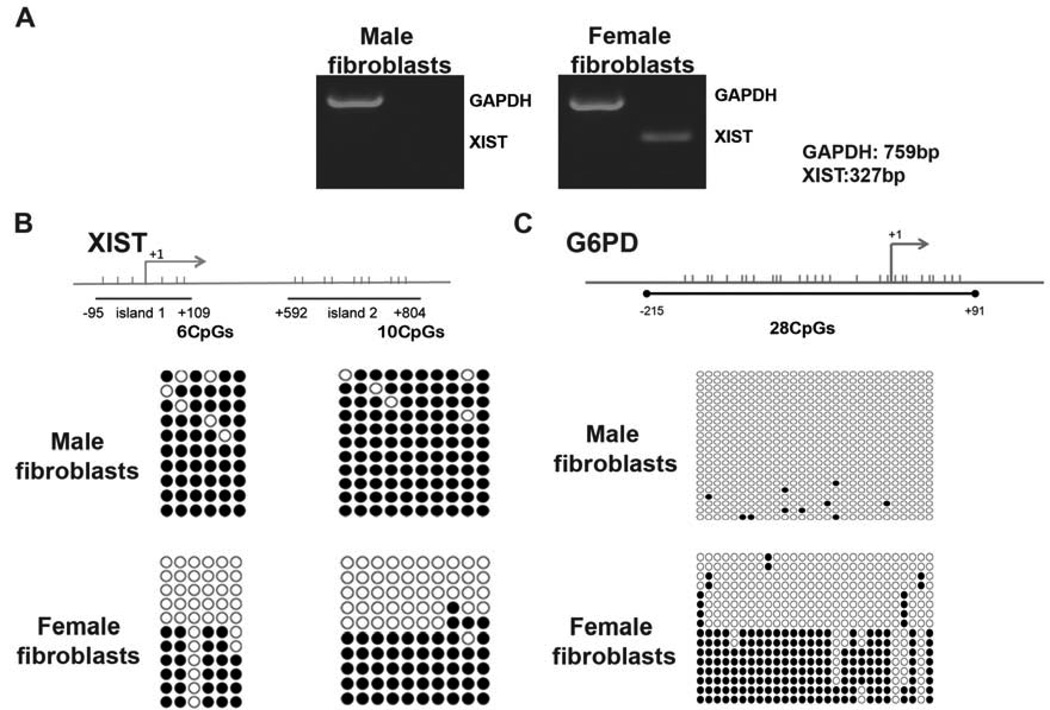
Figure 1. Expression and methylation of X-linked genes in rhesus macaque somatic cells.
(A) Expression of XIST was detected by RT-PCR in female but not male fibroblasts. Housekeeping gene GAPDH was used as a control.
(B) Methylation analysis of XIST promoter region by bisulfite sequencing. Horizontal bars indicate position of individual CpG dinucleotides within each region. The islands 1 and 2 consisting of six and ten CpG sites, respectively, were fully methylated in male fibroblasts indicating transcriptional silencing of XIST. However, approximately half of the clones in female fibroblasts were unmethylated suggesting expression of XIST from one X chromosome.
(C) Methylation profile of X-linked G6PD in fibroblasts. A total of twenty-eight CpG sites were analysed that were unmethylated in male samples in agreement with conclusion that males possess single but active X. As expected, half of the clones in female samples were methylated suggesting that one X chromosome is silenced.
Since expression of X-linked genes in a mixed population of female tissues displaying random XCI (XiXa+XaXi) would be similar to a scenario in which cells possess two active X chromosomes (XaXa), we explored DNA methylation as an additional epigenetic marker of XCI. It is generally accepted that XIST promoter region harboring CpG region is methylated on a silent XIST but otherwise transcriptionally active X chromosome (Norris et al., 1994). In contrast, this region is unmethylated on the opposite allele resulting in the transcription of XIST but silencing of other X-linked genes on this chromosome (Beard et al., 1995; Norris et al., 1994). We carried out methylation analysis of XIST by bisulfite genomic sequencing and focused on the two CpG islands located within promoter/exon 1, based on previously reported human studies (Hendrich et al., 1997; Poplinski et al., 2010; Shen et al., 2008). Information for primers for bisulfite sequencing and PCR conditions are presented in supplemental information (Table S4). Male somatic tissues displayed nearly complete methylation in both CpG islands, in agreement with the transcriptional silencing of XIST but otherwise active single X. In female tissues, approximately 50% of analysed clones were methylated for both islands (Fig. 1B). These results support the conclusion that methylation status of XIST correlates with its expression and can be used to differentiate random XCI from two active X chromosomes.
We also extended our methylation studies to G6PD, the gene also known to undergo XCI (Carrel and Willard, 2005; Keohane et al., 1996). In contrast to XIST, methylation of most X-linked genes is associated with their transcriptional silencing on the chromosome they reside on. As expected, male tissues were completely unmethylated within G6PD, consistent with the notion that male cells possess single but active X chromosome (Fig. 1C). In contrast, approximately half of the analysed clones in female tissues exhibited heavy methylation, suggesting that female somatic cells have undergone random XCI and that in any given cell, the X-linked genes are transcribed from one active X chromosome.
XCI in primate blastocysts
Next we explored XCI in monkey blastocysts produced in vitro using our conventional approaches (Wolf et al., 2004). We analysed XIST expression, separately, in the ICM and TE isolated from various stage blastocysts by immunosurgery or laser-assisted biopsy. We also used a portion of biopsied TE samples to differentiate male and female blastocysts using PCR-based gender analysis based on size differences in the amplicons of the X- and Y-linked zinc finger protein genes (ZFX and ZFY) (Mitalipov et al., 2007). Conventional RT-PCR analysis revealed the presence of XIST transcripts in all female ICMs (Fig. 2A). Interestingly, we also detected XIST expression in 4 out of 5 tested male ICMs, which was in agreement with previous observations in human embryos (Okamoto et al., 2011; Ray et al., 1997). Next, we conducted quantitative real-time (q)PCR analysis to determine levels of XIST expression during developmental progression of monkey blastocysts. In female ICMs, XIST expression levels gradually increased from early blastocysts (EB, day 6 post fertilization) to expanded blastocysts (ExB, day 8) and hatched blastocysts (HB, day 11). In contrast, male ICMs exhibited low levels of XIST that did not change during progression through blastulation stages (Fig. 2A). Female TE cells were assayed for expanded blastocysts only and displayed significant levels of XIST expression similar to that seen for ICMs in hatched blastocysts.
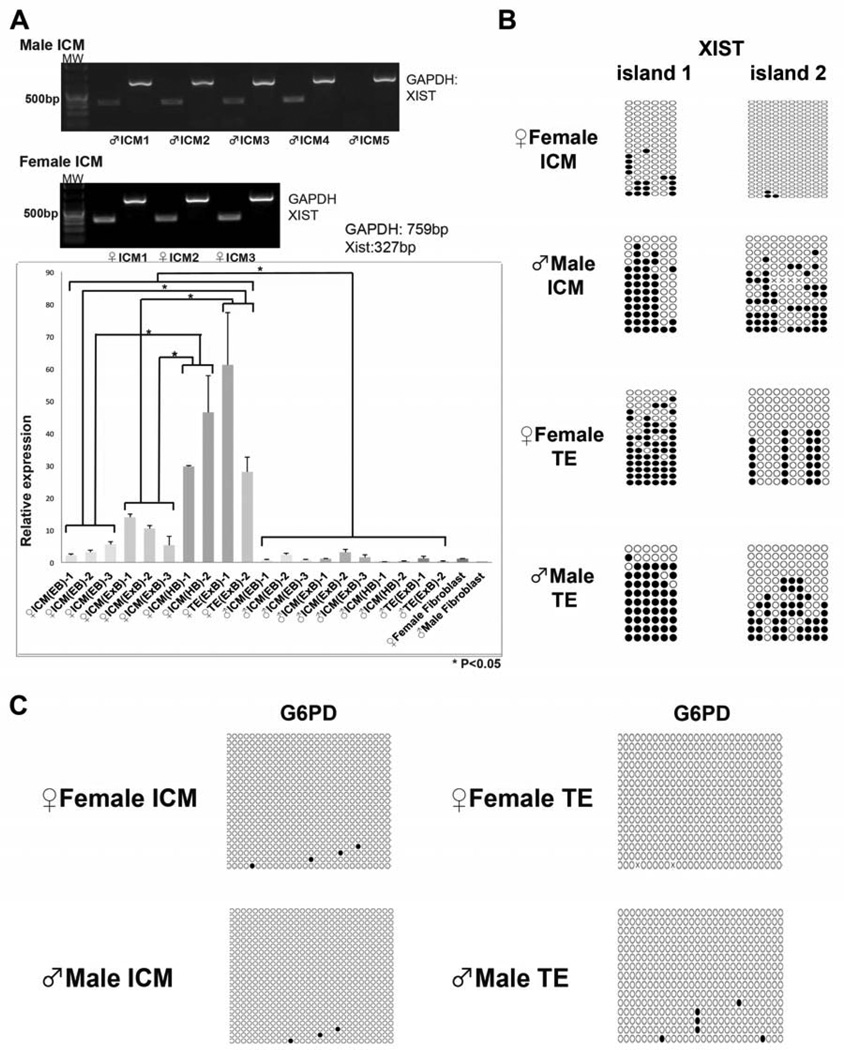
Figure 2. Expression and methylation of XIST and G6PD in monkey ICM and TE samples.
(A) XIST expression in isolated ICMs was analysed by qualitative and quantitative PCR. RT-PCR detected strong XIST expression in all female ICM samples while in most male ICMs expression was moderate. Levels of XIST expression were also analysed by qPCR. In female ICMs, XIST expression levels increased with developmental progression from early (EB) to expanded (ExB) and hatched blastocysts (HB). Male ICMs exhibited low levels of XIST that did not change throughout blastulation. Similar to ICMs, female TE cells also expressed significant levels of XIST. Abbreviations: MW, MF, FF, neg, EB, ExB, HB, ICM and TE indicate Molecular Weight, Male Fibroblasts, Female Fibroblasts, negative control, Early Blastocysts, Expanded Blastocysts, Hatched Blastocysts, Inner Cell Mass and Trophectoderm, respectively.
(B) XIST methylation profiles in the ICM and TE. In female ICMs few clones in the island 1 were sporadically methylated and the island 2 was completely unmethylated. In contrast, more than half of the clones were methylated in male ICMs and both gender TE samples.
(C) G6PD promoter was unmethylated in both male and female samples.
To further define XCI status in the ICM and TE of expanded blastocysts, we carried out methylation studies of XIST and G6PD as described above. Due to insufficient DNA amounts for bisulfite sequencing, up to twenty individual ICMs or three TEs were pooled for each assay. Both XIST CpG islands were hypomethylated in female ICMs (Fig. 2B), consistent with biallelic expression of XIST. In contrast, majority of clones in male ICMs showed sporadic methylation patterns (Fig. 2B). Levels of XIST methylation in male ICMs were notably lower than that seen in male somatic cells. This observation correlates with low but detectable XIST expression in male ICMs. XIST methylation patterns were comparable for both male and female TE samples, with heavy methylation in most clones for the island 1 while approximately half clones in the island 2 were sporadically methylated (Fig. 2B). In contrast, G6PD gene was unmethylated in the ICM and TE cells of both in female and male embryos, suggesting that this gene is transcribed from both X chromosomes (Fig. 2C).
We also carried out allele-specific expression analysis of XIST, ZNF41 and FMR-1 in individual ICMs and TEs based on SNPs identified in sperm and oocyte donors. We collected gametes from animals carrying informative alleles and generated heterozygous embryos as described previously (Wolf et al., 2004). As expected, male ICMs expressed these genes from the maternal allele and transcripts from both X chromosomes were detected in female ICMs (Table 1 and Fig. S1). Similar to the ICM, both maternal and paternal transcripts of XIST and ZNF41 were detected in TE samples, while FMR1 was expressed monoallelically from the paternal allele (Table 1). We also recovered placental tissues from two midgestation rhesus pregnancies and confirmed biallelic expression of XIST and ZNF41 (Table 1).
Table 1.
Allele-specific expression analysis of XIST, ZNF41 and FMR1 in the ICM, TE and placenta
| Gene | SNP * | ICM #1 | ICM #2 | ICM #3 | ICM #4 | ICM #5 | TE #1 | TE #2 | Placenta #1 |
Placenta #2 |
|---|---|---|---|---|---|---|---|---|---|---|
| XIST | A/G16344 | Bi m5/p9 |
Bi m3/p9 |
- | Bi m4/p10 |
|||||
| A/T 16396 | - | - | - | Bi m3/p9 |
Bi m3/p8 |
Bi m5/p9 |
Bi m3/p9 |
Bi m2/p13 |
- | |
| A/G 16479 | Bi m6/p4 |
Bi m9/p9 |
Bi m7/p9 |
- | - | - | - | Bi m2/p13 |
Bi m4/p10 |
|
| ZNF41 | C/T 3424 | Bi m2/p8 |
Bi m4/p8 |
Bi m1/p10 |
- | Bi m11/p3 |
Bi m1/p13 |
Bi m10/p5 |
- | - |
| A/G 3503 | Bi m2/p8 |
Bi m4/p8 |
Bi m1/p10 |
- | Bi m11/p3 |
Bi m1/p13 |
Bi m10/p5 |
- | - | |
| C/T 3535 | - | - | - | Bi m12/p1 |
- | - | - | Bi m10/p2 |
Bi m11/p1 |
|
| FMR1 | G/T 2623 | - | - | - | Bi m8/p6 |
- | Mono (p) |
- | - | - |
detailed information for each SNP position is presented in Supplementary Material, Table S2
“-” indicates absence of informative SNPs for this sample
Mono and Bi indicate monoallelic or biallelic expression, respectively
“m” and “p” indicates expression from the maternal and paternal alleles, respectively.
Number after “p” and “m” indicates number of individual clones expressing this particular parental allele.
In summary, these results indicate that XIST is expressed at significantly higher levels in female ICMs compared to male embryos. Based on the negative correlation between XIST methylation and expression seen in monkey somatic cells, lack of methylation marks suggests that XIST is expressed biallelically in female ICMs. We previously demonstrated that immunosurgically isolated monkey ICMs contain NANOG expressing epiblast and GATA-6 positive primitive endoderm cells (Tachibana et al., 2012). It is possible that XCI varies between these two different lineages in monkey ICMs.
Lack of G6PD methylation also supports the model that both X-chromosomes are transcriptionally active in monkey ICMs. In contrast, partial XIST methylation in the TE suggests that the process of XCI has been initiated in the TE lineage. However, allele-specific expression analysis of TE and placental samples suggests that XCI in the primate TE lineage is not strictly paternal.
XCI in cultured pluripotent stem cells
We next explored XCI status in several ESC lines derived from in vitro fertilized (IVF) blastocysts (ORMES lines) (Mitalipov et al., 2006). In addition, we studied experimental pluripotent stem cells derived by SCNT (CRES lines) (Byrne et al., 2007; Sparman et al., 2009). These cell lines were extensively characterized for expression of pluripotency markers and by in vitro and in vivo differentiation elsewhere (Byrne et al., 2007; Mitalipov et al., 2006; Sparman et al., 2009). We also generated monkey iPS cells (RiPS lines) from the same parental somatic cells that were used as nuclear donors for SCNT (Table S5) using retroviral transduction with vectors carrying OCT4, SOX2, Klf4 and cMYC. All RiPS cell lines were morphologically indistinguishable from their IVF- or SCNT-derived counterparts (Byrne et al., 2007; Sparman et al., 2009) and expressed conventional pluripotency markers including OCT4, NANOG, SSEA-4, TRA-1-60 and -1-81 (Fig. S2A). In addition, we conducted microarray analysis of genome-wide mRNA profiles of iPS cells in comparison to their IVF and SCNT controls using the Affymetrix Rhesus Macaque Genome array. Results confirmed that all monkey pluripotent stem cells exhibited similar transcriptional profiles (Fig. S2B).
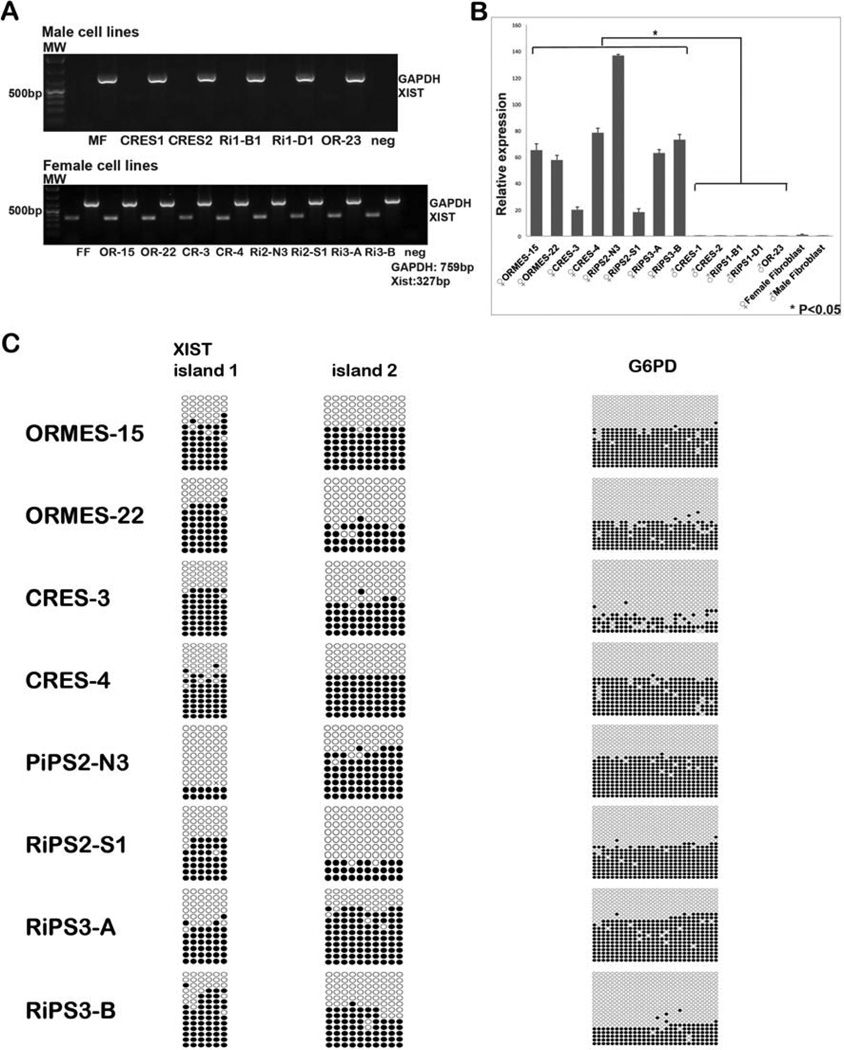
To exclude the possibility that XCI status may change during long-term culture, all stem cells were analysed at early passages. Similar to somatic cells, male pluripotent cells did not express XIST (Fig. 3A). However, female stem cells displayed significant levels of XIST transcripts comparable to that of female ICMs (Fig. 3B). We next conducted methylation analysis of stem cell lines by bisulfite sequencing. Approximately half clones in all female stem cell lines were heavily methylated in XIST and G6PD promoter regions (Fig. 3C and Fig. S3A). These results contrasted with methylation profiles seen in female ICMs but were similar to female somatic cells. To relate methylation with expression, we carried expression analysis of G6PD by qPCR (Fig. S3B) and observed that lack of methylation is associated with increased expression (Fig. S3C).
Figure 3. X-linked gene expression and methylation in rhesus macaque pluripotent stem cells.
(A) XIST expression by conventional RT-PCR was undetectable in male ESCs, whereas all female lines strongly expressed XIST similar to that seen in somatic cells. Abbreviations: MW, MF, FF, Ri1-B1, Ri1-D1, OR-23, OR-15, OR-22, CR-3, CR-4, Ri2-N3, Ri2-S1, Ri3-A, Ri3-B and neg indicate Molecular Weight, Male Fibroblasts, Female Fibroblasts, RiPS1-B1, RiPS1-D1, ORMES-23, ORMES-15, ORMES-22, CRES-3, CRES-4, RiPS2-N3, RiPS2-S1, RiPS3-A, RiPS3-B and negative control.
(B) qPCR based assay demonstrated significant levels of XIST expression in all female cell lines.
(C) Approximately half of the clones in all female pluripotent stem cells were methylated in both XIST promoter islands. G6PD methylation in female stem cells was similar to XIST indicating XCI.
Allele-specific expression of XIST, ZNF41 and FMR1 demonstrated that all informative female cell lines, regardless of their origin, expressed these genes from a single X-chromosome (Table 2). In two IVF-derived cell lines (ORMES-15 and 22), XIST expression was from the maternal X chromosome, while ZNF41 and FMR1 were transcribed from the opposite paternal X. Similar reciprocal expression pattern was also seen in CRES and iPS cell lines, although we were unable to determine parent-of-origin X-chromosomes in these cells since parental information for adult animals that contributed skin fibroblasts was unavailable. Interestingly, CRES-3, CRES-4, RiPS-3A and RiPS-3B were all derived from the same parental skin fibroblasts established from the 8 years old female #4 (Table S5). CRES-3, CRES-4 and RiPS-3A reciprocally expressed these genes from the same X-chromosomes. However, expression of XIST, ZNF41 and FMR1 in RIPS-3B was from opposite X chromosomes (Table 2).
Table 2.
Allele-specific expression of XIST, ZNF41 and FMR1 in monkey pluripotent stem cell lines
| Gene | SNP * | OR-15 | OR-22 | CRES-3 | CRES-4 | RiPS2- N3 |
RiPS2- S1 |
RiPS3- A |
RiPS3- B |
|---|---|---|---|---|---|---|---|---|---|
| XIST | C/T 13501 | - | - | - | - | Mono T |
Mono C |
- | - |
| A/G 15439 | - | Mono (m) |
- | - | - | - | - | - | |
| C/T 15491 | Mono (m) |
- | - | - | - | - | - | - | |
| C/T 15631 | Mono (m) |
- | - | - | - | - | - | - | |
| A/G 16344 | - | Mono (m) |
Mono G |
Mono G |
- | - | Mono G |
Mono A |
|
| A/T 16396 | Mono (m) |
- | - | - | - | - | - | - | |
| A/G 16479 | Mono (m) |
Mono (m) |
Mono A |
Mono A |
- | - | Mono A |
Mono G |
|
| ZNF41 | A/T 1448 | - | Mono (p) |
- | - | Mono A |
Mono T |
- | - |
| A/G 2148 | - | - | - | - | - | - | - | - | |
| C/T 3424 | - | Mono (p) |
Mono T |
Mono T |
- | - | Mono T |
Mono C |
|
| A/G 3503 | Mono (p) |
Mono (p) |
Mono A |
Mono A |
- | - | Mono A |
Mono G |
|
| C/T 3535 | - | - | Mono C |
Mono C |
- | - | Mono C |
Mono T |
|
| A/G 4047 | - | - | - | - | - | - | - | - | |
| FMR1 | G/T 2623 | - | - | - | - | Mono G |
Mono T |
- | - |
detailed information on SNP position is presented in Supplementary Material, Table S2
"-" indicates absence of informative SNPs for this sample
Mono and Bi indicate monoallelic or biallelic expression, respectively.
(m) and (p) indicates expression from the maternal or paternal alleles, respectively.
Parental information was only available for IVF-derived OR-15 and OR-22 cell lines
Collectively, these results suggest that cultured monkey pluripotent stem cells, irrespective of their origin, underwent XCI. However, in contrast to somatic cells with random XCI, each stem cell line represents a population of cells expressing X-linked genes from a single chromosome.
Discussion
Proper X-inactivation is critical for normal development of female organisms as well as for ESC differentiation and their utility in regenerative medicine. Yet, a number of unresolved questions remain regarding XCI in human preimplantation embryos and ESCs. We provide here a comprehensive analysis of XCI in monkey blastocysts and cultured pluripotent stem cells based on methylation and allele-specific expression of XIST and other X-linked genes. Prior studies suggested that RNA FISH assay for XIST, a gold-standard approach that is primarily used in the mouse, might not be sufficient for validation of XCI in human embryos and ESCs (Minkovsky et al., 2012; Shen et al., 2008). We established that methylation of XIST and G6PD promoter regions in combination with allele-specific expression of X-linked genes can provide an alternative assay for determination of XCI.
Our results indicate that both X chromosomes are active in monkey ICMs, while even early passage female ESCs have undergone strictly monoallelic XCI (Fig. S4). These results are in agreement with reported studies on human ESC lines indicating that a majority of analysed cell lines have also undergone XCI (Shen et al., 2008; Silva et al., 2008).
Analysis of a large number of human ESCs also identified that some cell lines retain two active X chromosomes similar to that seen in the mouse (Silva et al., 2008). However, these cell lines are unstable and undergo XCI during culture while still remaining undifferentiated. Another class of human ESCs, apparently, maintains XaXa but is unable to initiate proper XCI after differentiation (Dvash et al., 2010). These observations were made based on XIST expression combined with FISH analysis or enrichment of histone 3 lysine 27 tri-methylation (H3K27me3). Thus, it is unclear if XIST and other X-linked genes maintain biallelic expression in these ESCs.
In contrast, stable mouse XX ESCs maintain two active X chromosomes that apparently display proper random XCI only after in vitro differentiation (Monkhorst et al., 2008; Payer and Lee, 2008). However, as pointed above, female ESC lines from most mouse strains are unstable and rapidly lose one of the X chromosomes at early passages (Tesar, 2005). This phenomenon was noticed shortly after the first successful derivation of mouse ESCs, more than two decades ago (Nichols et al., 1990; Robertson et al., 1983). It remains unclear if chromosomal instability in mouse female ESCs is correlated with the onset of XCI. Developmental studies indicated that such aneuploidy compromises ability of ESCs to generate chimeras and subsequently germ-line transmission (Robertson et al., 1983; Zvetkova et al., 2005).
Knowledge of XCI in human embryos remains limited despite its importance for understanding early development and disease. However, mechanisms regulating XCI during early human embryo development appears to be different from that in mouse. For instance, recent FISH-based study reported that both the ICM and TE of human blastocysts carry active X chromosomes despite being coated with XIST RNA (Okamoto et al., 2011). Our result in the monkey blastocysts are in agreement with these observations and demonstrate that XIST and other X-linked genes are actively transcribed from both alleles in ICMs and TEs. It also appears that unlike mouse, there is no imprinted paternal XCI in the monkey TE lineage supporting prior human observations (Fan and Tran, 2011; Okamoto et al., 2011). Previous studies also showed that global demethylation in mutant mice does not interfere with proper XCI suggesting that other mechanisms may also be involved (Sado et al., 2000; Sado et al., 2004).
Monkey ESCs derived from these ICMs display XCI even at early passages suggesting that XCI occurs during the derivation of ESCs. Moreover, we demonstrate a strictly monoallelic expression of XIST and other X-linked genes in all tested monkey ESCs suggesting that XCI in these cell lines is not random. Novel human XaXa ESCs were successfully derived and maintained under optimal 5% oxygen concentrations mimicking in vivo conditions (Lengner et al., 2010). XIST expression was not detectable in these ESCs and SNP-chip assay revealed biallelic expression of X-linked genes. However, we routinely culture monkey embryos and ESCs at 5% oxygen and it seems that these conditions do not prevent XCI in monkey ESCs. It should be noted that culture medium for human ESCs was different than that used for monkey cells in this study. While our medium contained fetal bovine serum, human ESC culture medium used LIF and serum replacement (Lengner et al., 2010).
Mouse pluripotent stem cells were also derived from the epiblast of post-implantation embryos (EpiSCs) (Brons et al., 2007; Tesar et al., 2007). Interestingly, female EpiSCs display random XCI but apparently karyotypically are more stable than female ESCs. Despite their ability to differentiate in vitro or in vivo into teratomas, EpiSCs display limited ability to contribute to chimeras (Guo et al., 2009; Tesar et al., 2007). Recent studies have shown that EpiSCs can be reprogrammed into a more potent (“naive”) ESC state (Guo et al., 2009). Based on similarities in XCI and growth requirements, it was suggested that human ESCs most likely represent epiblast stem cells and naive state can be restored in human ESCs under conditions developed for the mouse (Hanna et al., 2010). Particularly, it was demonstrated that transduction of human ESCs with vectors carrying KLF4 and OCT4 and culture with inhibitors of glycogen synthase kinase 3β (GSK3β) and mitogen-activated protein kinase (ERK1/2) can reactivate a silent X chromosomes resulting in ESCs with XaXa (Hanna et al., 2010). However, long-term maintenance of XaXa in these ESCs appears to depend on constitutive overexpression of these factors (Buecker et al., 2010; Hanna et al., 2010).
XCI in monkey pluripotent cells derived using SCNT or iPS-based reprogramming resemble situation with IVF-derived ESCs since all analysed cell lines showed monoallelic XCI. It remains unclear whether monkey SCNT embryos temporarily reactivate X in the ICM. In iPS cell lines, monoallelic XCI could simply reflect the origin from a single somatic cell (Mekhoubad et al., 2012). This is also supported by observation that two iPS cell clones generated from the same parental fibroblasts displayed opposite XCI. This allows generation of genetically-identical iPS cells but expressing different set of X-linked genes similar to that observed with human iPS cells derived from patients with Rett syndrome (Amenduni et al., 2011). Long-term culture of pluripotent stem cells may also induce epigenetic changes affecting maintenance XCI (Dvash et al., 2010; Mekhoubad et al., 2012; Shen et al., 2008; Silva et al., 2008).
In summary, we demonstrate the fundamental epigenetic differences between primate ex vivo pluripotent cells residing in blastocysts and their in vitro cultured counterparts. How XCI may affect human or monkey ESC differentiation and engraftment after transplantation remains unclear. Particularly, given that XCI in primate ESCs and iPS cells is non-random, rendering cells expressing X-linked genes from only one X chromosome. It is likely that similar to the mouse, X inactivation status in monkey ESCs affects their developmental potential. Thus, it is critical to understand mechanisms of XCI and to develop optimized culture conditions that would favour derivation and maintenance of ESCs with two active X-chromosomes.
Supplementary Material
Highlights.
-
●
Monkey Inner Cell Mass maintain two active X chromosomes
-
●
Monkey trophectoderm cells express X-linked genes from both alleles
-
●
ESCs display monoallelic X-inactivation
Acknowledgments
The authors would like to acknowledge the Division of Animal Resources, Surgery Team, Assisted Reproductive Technology & Embryonic Stem Cell Core, Endocrine Technology Core, and Molecular & Cellular Biology Core at the Oregon National Primate Research Center for providing expertise and services that contributed to this project. We are grateful to Lisa Clepper and Maidina Tuohetahuntila for their technical support. This study was supported by start up funds from Oregon National Primate Research Center and grants from the National Institutes of Health HD057121, HD059946, HD063276, EY021214, HD018185 and RR000163.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no financial affiliation that may be perceived as biasing this work.
The authors indicate no potential conflicts of interest.
SUPPLEMENTARY MATERIAL
Supplementary Materials include four figures and five tables.
REFERENCES
- Amenduni M, De Filippis R, Cheung AY, Disciglio V, Epistolato MC, Ariani F, Mari F, Mencarelli MA, Hayek Y, Renieri A, Ellis J, Meloni I. iPS cells to model CDKL5-related disorders. European journal of human genetics : EJHG. 2011;19:1246–1255. doi: 10.1038/ejhg.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buecker C, Chen HH, Polo JM, Daheron L, Bu L, Barakat TS, Okwieka P, Porter A, Gribnau J, Hochedlinger K, Geijsen N. A murine ESC-like state facilitates transgenesis and homologous recombination in human pluripotent stem cells. Cell Stem Cell. 2010;6:535–546. doi: 10.1016/j.stem.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, Wolf DP, Mitalipov SM. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Diaz-Perez S, Ouyang Y, Perez V, Cisneros R, Regelson M, Marahrens Y. The element(s) at the nontranscribed Xist locus of the active X chromosome controls chromosomal replication timing in the mouse. Genetics. 2005;171:663–672. doi: 10.1534/genetics.105.043026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvash T, Lavon N, Fan G. Variations of X chromosome inactivation occur in early passages of female human embryonic stem cells. PLoS One. 2010;5:e11330. doi: 10.1371/journal.pone.0011330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fan G, Tran J. X chromosome inactivation in human and mouse pluripotent stem cells. Human genetics. 2011;130:217–222. doi: 10.1007/s00439-011-1038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto A, Mitalipov SM, Kuo HC, Wolf DP. Aberrant genomic imprinting in rhesus monkey embryonic stem cells. Stem Cells. 2006;24:595–603. doi: 10.1634/stemcells.2005-0301. [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Surani MA. Development. Programming the X chromosome. Science. 2004;303:633–634. doi: 10.1126/science.1094408. [DOI] [PubMed] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich BD, Plenge RM, Willard HF. Identification and characterization of the human XIST gene promoter: implications for models of X chromosome inactivation. Nucleic Acids Res. 1997;25:2661–2671. doi: 10.1093/nar/25.13.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Keohane AM, O'Neill L P, Belyaev ND, Lavender JS, Turner BM. X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, Santagata S, Powers D, Barrett CB, Young RA, Lee JT, Jaenisch R, Mitalipova M. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. Erosion of Dosage Compensation Impacts Human iPSC Disease Modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkovsky A, Patel S, Plath K. Concise review: Pluripotency and the transcriptional inactivation of the female Mammalian X chromosome. Stem Cells. 2012;30:48–54. doi: 10.1002/stem.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitalipov S, Clepper L, Sritanaudomchai H, Fujimoto A, Wolf D. Methylation status of imprinting centers for H19/IGF2 and SNURF/SNRPN in primate embryonic stem cells. Stem Cells. 2007;25:581–588. doi: 10.1634/stemcells.2006-0120. [DOI] [PubMed] [Google Scholar]
- Mitalipov S, Kuo HC, Byrne J, Clepper L, Meisner L, Johnson J, Zeier R, Wolf D. Isolation and characterization of novel rhesus monkey embryonic stem cell lines. Stem Cells. 2006;24:2177–2186. doi: 10.1634/stemcells.2006-0125. [DOI] [PubMed] [Google Scholar]
- Monkhorst K, Jonkers I, Rentmeester E, Grosveld F, Gribnau J. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell. 2008;132:410–421. doi: 10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- Moreira de Mello JC, de Araujo ES, Stabellini R, Fraga AM, de Souza JE, Sumita DR, Camargo AA, Pereira LV. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS One. 2010;5:e10947. doi: 10.1371/journal.pone.0010947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Evans EP, Smith AG. Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development. 1990;110:1341–1348. doi: 10.1242/dev.110.4.1341. [DOI] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Norris DP, Patel D, Kay GF, Penny GD, Brockdorff N, Sheardown SA, Rastan S. Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell. 1994;77:41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf JP, Renard JP, Duranthon V, Heard E. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- Panning B, Dausman J, Jaenisch R. X chromosome inactivation is mediated by Xist RNA stabilization. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annual review of genetics. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Poplinski A, Wieacker P, Kliesch S, Gromoll J. Severe XIST hypomethylation clearly distinguishes (SRY+) 46,XX-maleness from Klinefelter syndrome. European journal of endocrinology / European Federation of Endocrine Societies. 2010;162:169–175. doi: 10.1530/EJE-09-0768. [DOI] [PubMed] [Google Scholar]
- Ray PF, Winston RM, Handyside AH. XIST expression from the maternal X chromosome in human male preimplantation embryos at the blastocyst stage. Human molecular genetics. 1997;6:1323–1327. doi: 10.1093/hmg/6.8.1323. [DOI] [PubMed] [Google Scholar]
- Robertson EJ, Evans MJ, Kaufman MH. X-chromosome instability in pluripotential stem cell lines derived from parthenogenetic embryos. Journal of embryology and experimental morphology. 1983;74:297–309. [PubMed] [Google Scholar]
- Sado T, Fenner MH, Tan SS, Tam P, Shioda T, Li E. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Developmental biology. 2000;225:294–303. doi: 10.1006/dbio.2000.9823. [DOI] [PubMed] [Google Scholar]
- Sado T, Okano M, Li E, Sasaki H. De novo DNA methylation is dispensable for the initiation and propagation of X chromosome inactivation. Development. 2004;131:975–982. doi: 10.1242/dev.00995. [DOI] [PubMed] [Google Scholar]
- Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD, Fan G. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci U S A. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparman M, Dighe V, Sritanaudomchai H, Ma H, Ramsey C, Pedersen D, Clepper L, Nighot P, Wolf D, Hennebold J, Mitalipov S. Epigenetic reprogramming by somatic cell nuclear transfer in primates. Stem Cells. 2009;27:1255–1264. doi: 10.1002/stem.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sritanaudomchai H, Ma H, Clepper L, Gokhale S, Bogan R, Hennebold J, Wolf D, Mitalipov S. Discovery of a novel imprinted gene by transcriptional analysis of parthenogenetic embryonic stem cells. Human reproduction. 2010;25:1927–1941. doi: 10.1093/humrep/deq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sparman M, Ramsey C, Ma H, Lee HS, Penedo MC, Mitalipov S. Generation of chimeric rhesus monkeys. Cell. 2012;148:285–295. doi: 10.1016/j.cell.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sparman M, Sritanaudomchai H, Ma H, Clepper L, Woodward J, Li Y, Ramsey C, Kolotushkina O, Mitalipov S. Mitochondrial gene replacement in primate offspring and embryonic stem cells. Nature. 2009;461:367–372. doi: 10.1038/nature08368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tesar PJ. Derivation of germ-line-competent embryonic stem cell line s from prebla stocyst mouse embryos. Proc Natl Acad Sci U S A. 2005;102:8239–8244. doi: 10.1073/pnas.0503231102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP, Becker RA, Hearn JP. Isolation of a primate embryonic stem cell line. Proc Natl Acad Sci U S A. 1995;92:7844–7848. doi: 10.1073/pnas.92.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg IM, Laven JS, Stevens M, Jonkers I, Galjaard RJ, Gribnau J, van Doorninck JH. X chromosome inactivation is initiated in human preimplantation embryos. Am J Hum Genet. 2009;84:771–779. doi: 10.1016/j.ajhg.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DP, Thormahlen S, Ramsey C, Yeoman RR, Fanton J, Mitalipov S. Use of assisted reproductive technologies in the propagation of rhesus macaque offspring. Biol Reprod. 2004;71:486–493. doi: 10.1095/biolreprod.103.025932. [DOI] [PubMed] [Google Scholar]
- Wu D, Hamilton B, Martin C, Gao Y, Ye M, Yao S. Generation of induced pluripotent stem cells by reprogramming human fibroblasts with the stemgent human TF lentivirus set. J Vis Exp. 2009 doi: 10.3791/1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng SM, Yankowitz J. X-inactivation patterns in human embryonic and extra-embryonic tissues. Placenta. 2003;24:270–275. doi: 10.1053/plac.2002.0889. [DOI] [PubMed] [Google Scholar]
- Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, John R, Feil R, Brockdorff N. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.