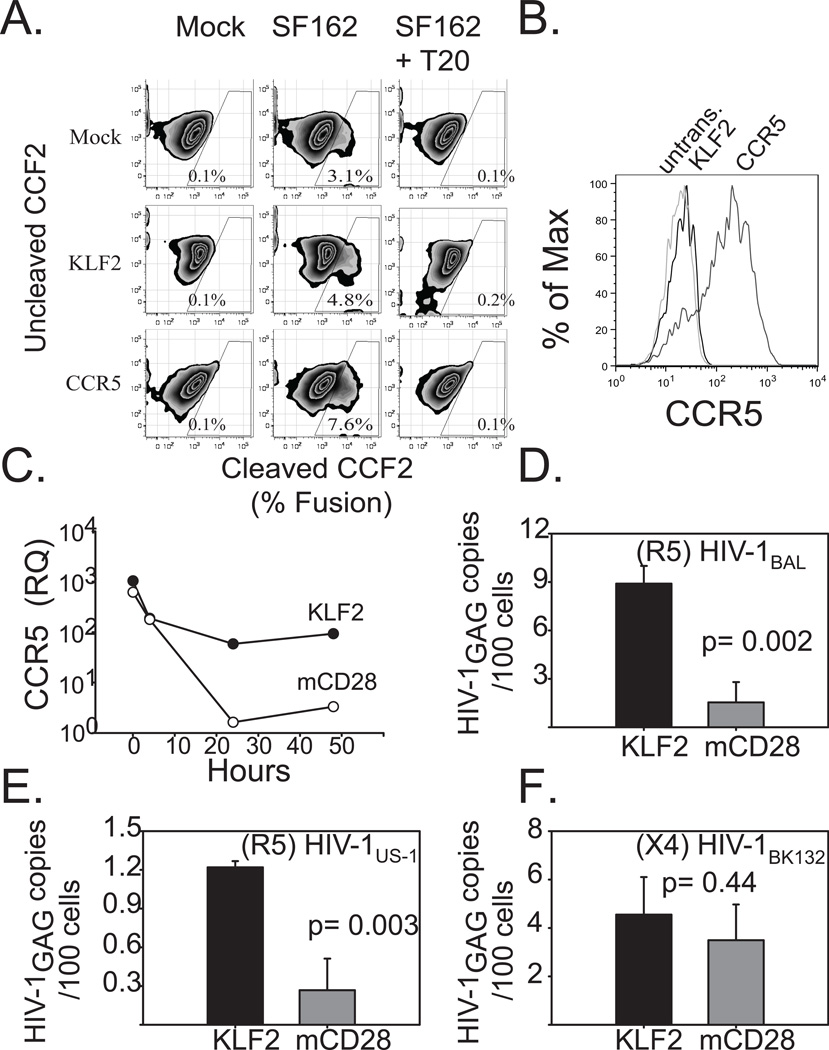
Figure 6. Forced expression of KLF2 in CD4s restores CCR5 expression during CD3/28 costimulation, and renders them susceptible to R5-tropic HIV.
A. PHA + IL2 stimulated primary human CD4 T cells were transduced with mRFP 2A KLF2-, or CCR5-expressing lentiviral vectors, or were left untransduced as a control. After 4 days of culture, KLF2- expressing cells were sorted based on mRFP expression. The following day cells were loaded with CCF2 and challenged with HIV-1 pseudotype expressing the HIVSF162 (R5) Env and Vpr-BlaM fusion in the presence or absence of 1μg of T20. Cleaved CCF2 was analyzed by flow cytometry 24 hours later. B. Untransduced and KLF2-, and CCR5-expressing CD4 T cells used in the BlaM assay were stained for cell surface CCR5 expression and assayed via flow cytometry. C. Untransduced, sorted KLF2-, or sorted mCD28-167*- expressing human CD4 T cells were activated with αCD3/CD28-coated beads. Relative CCR5 expression levels were determined by real time prior to (T = 0 hrs) and during αCD3/CD28 coated bead costimulation (T = 4, 24, 48 hrs). Expression levels were normalized to those in untransduced, αCD3/CD28 costimulated CD4 T cells at assay endpoint (T = 48 hrs). Similar results were obtained in five separate experiments, using cells from different donors. D,E.F KLF2- and mCD28-167*- expressing human CD4 T cells were prepared as above and activated with αCD3/CD28-coated beads and then challenged two hours later with CCR5-tropic or CXCR4-tropic cell-free HIV-1. After 48 hours, DNA was isolated from harvested cells, and real time PCR was performed for integrated HIV-1 gag DNA. DNA from stably-infected Ach-2 cells was used as a standard. D. CCR5-tropic HIV-1BaL challenge. E. CCR5-tropic HIV-1US1 challenge. F. CXCR4-tropic HIV-1BK132 challenge. Each experiment was performed in duplicate and data shown is representative of three independent studies. Error bars reflect the standard deviation of real time PCR assay of one representative experiment.