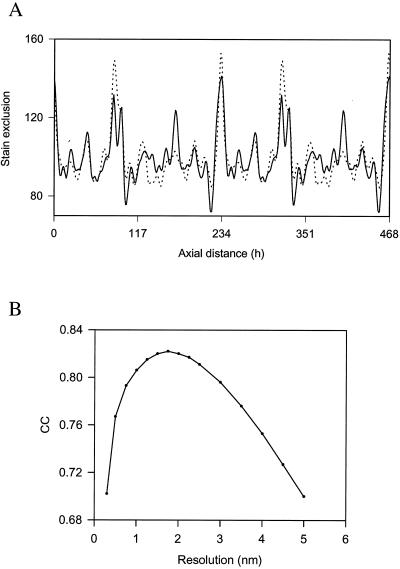
Figure 2.
Resolution in the axial structure. (A) Comparison of a theoretical (dotted line) stain-exclusion pattern with experimental (solid line) data from interior 4-nm-thick slices of the three-dimensional reconstruction, averaged over 20 D-periods. The theoretical pattern is based on the known axial structure of the triple helix and established correlation between stain exclusion and “bulkiness” (= volume/chain length) of each amino acid residue (43). The gap structure has been modeled in a compact form thereby removing the gap–overlap contrast in the fibril interior. The contributions of the N- and C-telopeptides have been calculated by using contraction factors, with respect to the residue spacing in the triple helix, of 0.3 and 0.7, respectively. The theoretical pattern has been smoothed to a resolution of 18 Å. h, axial residue spacing where D (67 nm) = 234 residues. (B) Plot of cross correlation coefficient (CC) between the experimental stain-exclusion pattern and the resolution of the smoothed theoretical pattern.