Abstract
The first step of nitrification is catalysed by both ammonia-oxidizing bacteria (AOB) and archaea (AOA), but physicochemical controls on the relative abundance and function of these two groups are not yet fully understood, especially in freshwater environments. This study investigated ammonia-oxidizing populations in nitrifying rotating biological contactors (RBCs) from a municipal wastewater treatment plant. Individual RBC stages are arranged in series, with nitrification at each stage creating an ammonia gradient along the flowpath. This RBC system provides a valuable experimental system for testing the hypothesis that ammonia concentration determines the relative abundance of AOA and AOB. The results demonstrate that AOA increased as ammonium decreased across the RBC flowpath, as indicated by qPCR for thaumarchaeal amoA and 16S rRNA genes, and core lipid (CL) and intact polar lipid (IPL) crenarchaeol abundances. Overall, there was a negative logarithmic relationship (R2 = 0.51) between ammonium concentration and the relative abundance of AOA amoA genes. A single AOA population was detected in the RBC biofilms; this phylotype shared low amoA and 16S rRNA gene homology with existing AOA cultures and enrichments. These results provide evidence that ammonia availability influences the relative abundances of AOA and AOB, and that AOA are abundant in some municipal wastewater treatment systems.
Introduction
Ammonia is a metabolic waste product that threatens aquatic ecosystems with toxicity, oxygen depletion and algal blooms. A primary objective of wastewater treatment is to prevent these adverse environmental impacts by removing ammonia from wastewater prior to discharge into receiving waters. Ammonia removal in wastewater treatment is accomplished through nitrification, a microbially mediated process in which ammonia is oxidized to nitrite and subsequently to nitrate. Wastewater treatment plants (WWTPs) may release nitrate-rich effluents, or nitrate may be reduced to dinitrogen gas via anaerobic ammonia oxidation (anammox) or denitrification prior to effluent discharge. Until recently, only specific members of the Beta- and Gammaproteobacteria, known as ammonia-oxidizing bacteria (AOB), were believed to be capable of catalysing the process of ammonia oxidation. Understanding of nitrification in natural environments changed with the discovery that members of the newly proposed phylum Thaumarchaeota (Brochier-Armanet et al., 2008; Spang et al., 2010) are capable of ammonia oxidation (Könneke et al., 2005), and that ammonia-oxidizing archaea (AOA) are ubiquitous in natural environments (Francis et al., 2005; Prosser and Nicol, 2008). In fact, AOA outnumber AOB in a variety of environments including soils (Leininger et al., 2006), marine habitats (Wuchter et al., 2006; Beman et al., 2008; De Corte et al., 2008) and estuarine sediments (Beman and Francis, 2006). However, AOB are numerically dominant in some sampled environments, including the San Francisco Bay Estuary (Mosier and Francis, 2008), as well as industrial (Limpiyakorn et al., 2010) and municipal (Wells et al., 2009) WWTPs.
Recent studies suggest that ammonia availability influences niche separation of AOA and AOB. Kinetic studies of the AOA isolate Nitrosopumilus maritimus demonstrate an exceptionally high substrate affinity for ammonia; N. maritimus has a half saturation constant (Km) of 0.133 µM total ammonia (Martens-Habbena et al., 2009), which is approximately two orders of magnitude lower than that of Nitrosomonas oligotropha, for which reported Km values range from 30 to 75 µM total ammonia (Stehr et al., 1995). As a result of this high substrate affinity andthe high relative abundance of AOA in oligotrophic environments, ammonia availability has been suggested to be an important factor in determining niche partitioning of AOA and AOB (Erguder et al., 2009; Schleper, 2010). Studies of soil AOA support this observation by demonstrating that AOB are numerically and metabolically dominant in ammonium-amended soils (Jia and Conrad, 2009; Di et al., 2010; Taylor et al., 2010; Verhamme et al., 2011). However, few studies have examined the role of ammonia availability in determining relative abundances of AOA and AOB in freshwater environments, especially those in engineered water treatment systems.
This study utilized existing nitrification infrastructure in a municipal WWTP in Guelph, Ontario, Canada to investigate the effect of ammonia concentrations on the abundance and diversity of prokaryotic ammonia-oxidizing communities. The Guelph WWTP features a tertiary treatment system of rotating biological contactors (RBCs), which are nitrification bioreactors comprised of panels of corrugated polymeric medium (Fig. 1A) attached to a central rotating shaft and partially submerged in wastewater. Eight individual RBC stages are arranged in series (Fig. 1B and C), with nitrification at each stage creating an ammonia gradient across the RBC flowpath. Although previous studies have detected AOB in nitrifying RBCs through PCR-based methods and microscopy (Egli et al., 2003; Pynaert et al., 2003; Jang et al., 2005), no study has investigated the occurrence, abundance or diversity of AOA in nitrifying RBCs. We hypothesized that the abundance of AOA would increase across the RBC flowpath, as wastewater becomes increasingly depleted of ammonium.
Fig. 1.
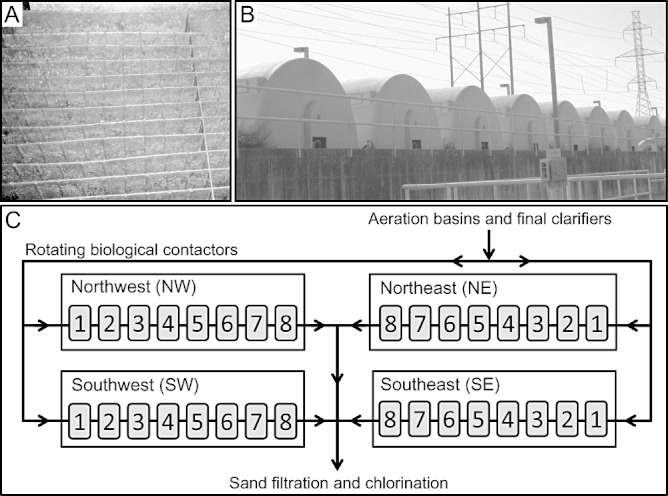
Outline of the sampling site in Guelph, Ontario. Internal medium of an RBC (A), an external view of a full RBC treatment train (B) and a schematic of the RBC arrangement in the Guelph WWTP (C).
Results
Water chemistry
In all sampling seasons and both the Northeast (NE) and Southeast (SE) treatment trains (see Experimental procedures for sampling details), ammonium decreased alongthe RBC flowpaths (Fig. 2A and Table S1). Overall, wastewater ammonium concentrations were highest in February and lowest in September, and were consistently higher in the NE treatment train than the SE train. Nitrite decreased across RBC flowpaths in patterns similar to ammonium, and nitrite concentrations were always relatively low (i.e. < 400 µg l−1, Table S1). In contrast, nitrate concentrations were always high (15–30 mg l−1), and measured nitrate levels did not change in a predictable manner across individual RBC flowpaths. For all RBC stages in all seasons, the pH varied within a narrow range of 7.2 to 7.6. Other parameters, such as temperature and dissolved organic carbon (DOC), varied little across a given RBC flowpath but showed seasonal differences. Dissolved oxygen (dO2) in this aerated system was always greater than 6 mg l−1, and increased by ≤ 2 mg l−1 across a given RBC flowpath.
Fig. 2.
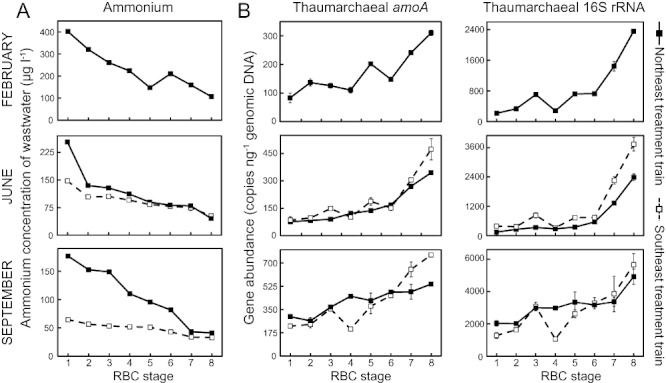
Ammonium concentrations of wastewater (A) and thaumarchaeal amoA and 16S rRNA gene abundances in associated biofilm samples (B) across RBC flowpaths. Error bars represent standard deviations based on technical duplicates; error bars that are not seen are contained within the symbols, except for ammonium concentrations in the February NE treatment train, for which duplicate measurements are not available.
Gene abundances
Thaumarchaeal amoA and 16S rRNA gene sequences were detected in all RBC stages from all seasons. In each RBC train sampled, AOA amoA gene abundance in genomic DNA extracts obtained from biofilm increased across the RBC flowpath (Fig. 2B). In addition, thaumarchaeal 16S rRNA gene abundance increased across the flowpath, in patterns congruent with archaeal amoA genes (Fig. 2B). For both thaumarchaeal amoA and 16S rRNA genes, abundance varied by season; gene abundances were highest in September and lowest in February (Fig. 2B). In addition, in both June and September, gene abundances were higher in the SE treatment train than the NE train.
Bacterial amoA genes were detected in biofilm extracts from all RBC stages. In contrast to AOA-associated genes, bacterial amoA gene abundance did not show predictable or consistent patterns between or across RBC trains when analysed independently (Fig. S1A). In addition, general bacterial 16S rRNA gene abundances (measured as a control gene) were consistent across all RBC stages, regardless of treatment train or season (Fig. S1B). When biofilm and associated wastewater from all RBC stages (i.e. from all sampling times and treatment trains) were considered together, the relative abundance of AOA amoA genes (as a proportion of total amoA genes) comprised 10–61% of the total ammonia-oxidizing community (Fig. 3). The relative abundance of AOA amoA genes demonstrated a negative logarithmic trend with ammonium concentration (R2 = 0.51; Fig. 3). Furthermore, Spearman's rank correlation coefficients were negative and statistically significant for influent ammonia concentrations with both the relative abundance of AOA amoA genes (i.e. as a proportion of total amoA genes; r = −0.6887, P < 0.0001) as well as independent AOA amoA gene abundances (r = −0.6088, P < 0.0001).
Fig. 3.
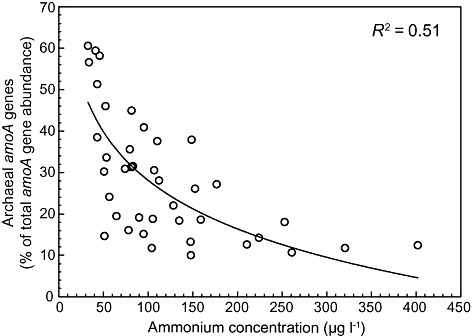
Ammonium concentrations of RBC-associated wastewater and relative abundance of archaeal amoA genes (as a per cent of total archaeal and bacterial amoA genes per nanogram of genomic DNA) in corresponding RBC biofilm.
Lipid abundances
In addition to quantification of key genes, concentrations of crenarchaeol, the characteristic membrane lipid of ammonia-oxidizing thaumarchaea (Pitcher et al., 2011a and references therein) were determined both as core lipids (CLs) as well as intact polar lipids (IPLs). Core lipids are assumed to represent fossilized (i.e. dead) biomass, while IPLs are biomarkers indicative of viable microbial cells (Sturt et al., 2004; Biddle et al., 2006; Pitcher et al., 2009; 2011b). Crenarchaeol in biofilm samples from all RBC stages of the June NE treatment train was quantified; both CL- and IPL-derived crenarchaeol increased across the RBC flowpath (Fig. 4A). For all other RBC trains, lipid analyses were performed for RBCs 1 and 8 only. In all cases, biofilm samples from RBC 8 yielded higher CL and IPL crenarchaeol abundances than RBC 1 (Fig. 4B).
Fig. 4.
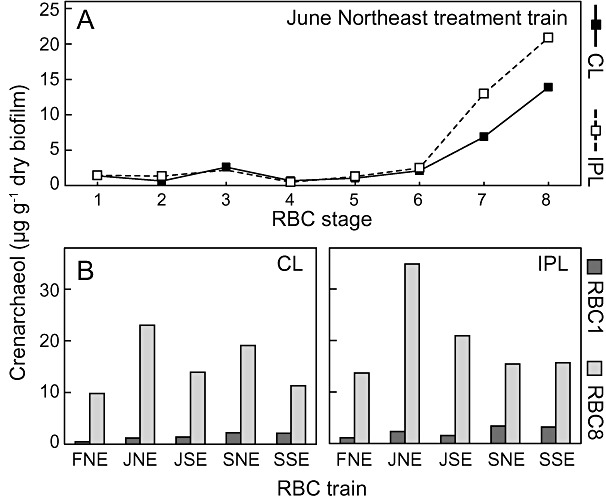
Lipid analysis. CL- and IPL-derived crenarchaeol abundance in biofilm across the June NE RBC flowpath (A) and core and IPL-derived crenarchaeol abundances for RBC 1 and 8 biofilm for all seasons and treatment trains (B). Letters F, J and S denote February, June and September respectively.
Gene diversity across RBC flowpath
For all genes analysed, communities were highly similar across all RBC stages of a given flowpath (Figs 5 and S2). Denaturing gradient gel electrophoresis (DGGE) profiles for thaumarchaeal amoA and 16S rRNA genes revealed simple patterns and low diversity; one dominant band was observed for both archaeal amoA and thaumarchaeal 16S rRNA (Fig. 5). For both thaumarchaeal genes analysed, diversity and community composition were nearly identical across all RBC stages sampled in all seasons (Figs 5 and S2). Bacterial amoA DGGE profiles were more complex, and patterns were similar across a given flowpath and between seasons (Figs 5 and S2). General bacterial 16S rRNA gene fingerprints were complex and highly similar, both across RBCs of a given treatment train (Fig. 5) and between treatment trains and seasons (Fig. S2).
Fig. 5.
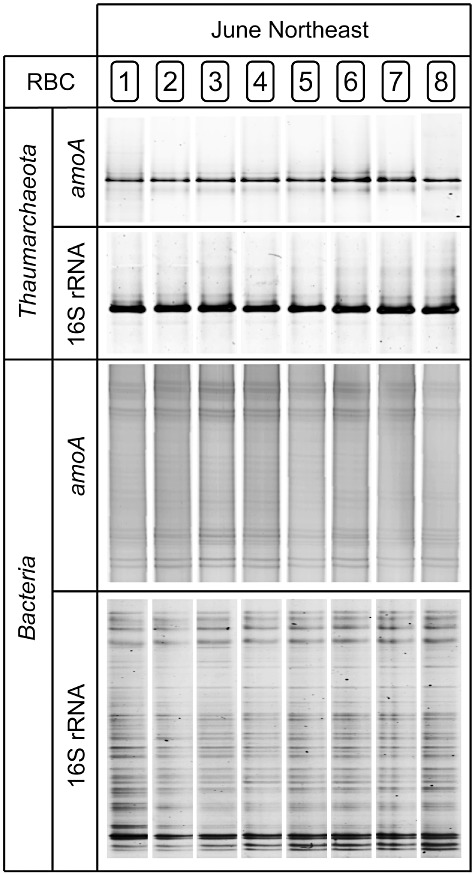
DGGE fingerprints for bacterial and thaumarchaeal amoA and 16S rRNA genes across eight serial RBC stages. Data shown were obtained from the NE treatment train, sampled in June 2010. Data from all RBC trains are shown in Fig. S1.
As described above, DGGE profiles showed simple patterns for archaeal amoA and 16S rRNA genes. Representative bands from both treatment trains (NE and SE) from all seasons were excised from gels and sequenced to confirm that populations were identical across all RBC stages, treatment trains and seasons. The resulting amoA sequence clustered with environmental sequences derived from other engineered environments, including landfills and activated sludge of municipal WWTPs (Fig. 6A). The amoA sequence retrieved from the Guelph RBCs shared a sequence identity of 94% with its most closely related sequences, which each originated from municipal WWTPs (HM589803, GU936642). Similarly, the thaumarchaeal 16S rRNA gene sequence obtained from Guelph WWTP clustered with genes retrieved from high-nutrient environments such as fertilized soils and WWTPs, and clustered distinctly from isolated or enriched AOA representatives (Fig. 6B). This sequence shared 100% sequence identity with sequences retrieved from an industrial WWTP (treating oil refinery waste) (HQ316970), activated sludge from a municipal WWTP (HM639792), and with sludge from a nitrous oxide oxidation system (AB619712).
Fig. 6.
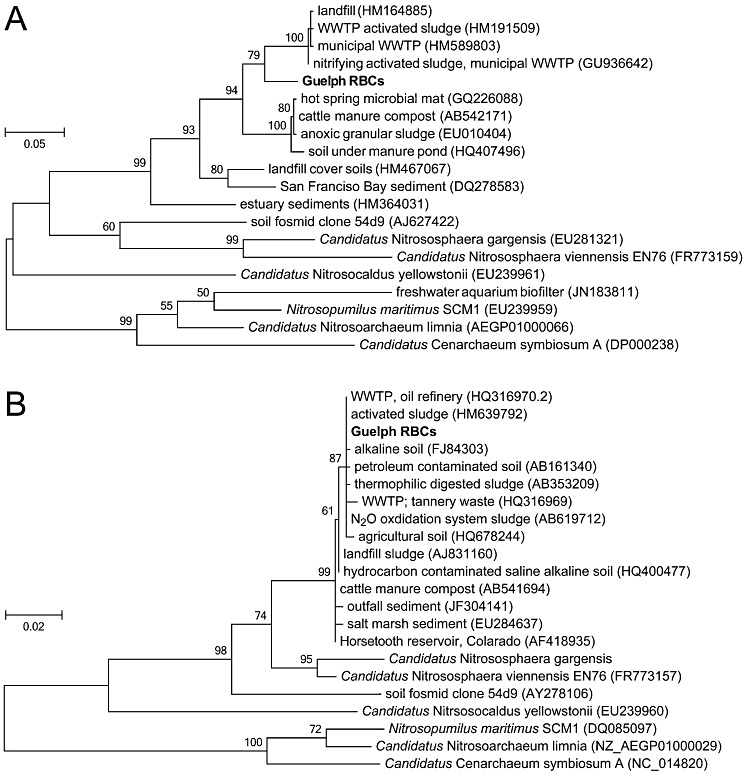
Phylogenetic affiliations of thaumarchaeal amoA (A) and 16S rRNA (B) gene sequences retrieved from Guelph WWTP RBCs. Both trees were inferred using the Maximum Likelihood method based on the Tamura–Nei model of sequence evolution. Bootstrap values are located above branches and are based on 500 replicates. Only bootstrap values greater than 50% are indicated on tree. The scale bars represent 5% and 2% nucleotide divergence for A and B respectively.
Discussion
The present study suggests niche adaptation of AOA to low-ammonia conditions, and identifies a wastewater environment in which AOA are abundant. For each RBC treatment train analysed, AOA populations increased in abundance as ammonia decreased along the flowpath (Fig. 2). When all RBCs from all seasons were analysed together, a negative correlation with high statistical significance (r = −0.6887, P < 0.0001) was observed between ammonium concentration of wastewater and relative abundance of AOA, implying that ammonia availability is an important factor in determining the relative proportions of ammonia-oxidizing populations.
Ammonium concentrations in the SE treatment train were consistently lower than in the NE treatment train, presumably because this train is located farther from the influent source (see Fig. 1C), and nitrification and volatilization may occur in the wastewater en route to the treatment train. Thaumarchaeal amoA and 16S rRNA genes were consistently more abundant in the SE treatment train (Fig. 2B), further supporting the inference of niche adaptation of AOA to low-ammonia conditions. Moreover, AOA gene abundances were highest in September, when ammonia concentrations were lowest. We expected AOB amoA genes to decrease across a given RBC flowpath in a pattern that was opposite to that observed for thaumarchaeal genes. However, AOB abundances showed no consistent pattern across the RBCs, indicating that ammonia concentrations remained in a range suitable for AOB growth. On the other hand, our data suggest that ammonia concentrations approached ranges suitable for the detected Thaumarchaeota towards RBC 8.
In all RBC stages from all seasons, AOA comprised a substantial proportion of the ammonia-oxidizing community (i.e. always greater than 10%), which is an important finding because studies of activated sludge in municipal WWTPs consistently show that AOB dominate ammonia-oxidizing communities, with AOA having low abundance or being absent (Park et al., 2006; Wells et al., 2009; Jin et al., 2010; Zhang et al., 2011). These studies have investigated suspended growth treatment systems used for secondary treatment, where high ammonia concentrations likely preclude the growth of AOA. Although two previous studies have suggested that AOA are abundant in aeration basins of municipal WWTPs (Limpiyakorn et al., 2010; Kayee et al., 2011), both studies used circular plasmids for qPCR standards, a practice that has been shown to cause serious overestimation of absolute gene copy numbers (Hou et al., 2010). Therefore, it remains unlikely that AOA are abundant in traditional aeration basin treatment systems.
Although overall abundance patterns between thaumarchaeal amoA and 16S rRNA genes were consistent (Fig. 2B), thaumarchaeal 16S rRNA genes were approximately an order of magnitude higher than archaeal amoA genes. It is currently unknown whether all Thaumarchaeota oxidize ammonia; however, two lines of evidence suggest that all thaumarchaeal populations in the Guelph WWTP RBCs possess amoA genes. First, the similarity between thaumarchaeal amoA and 16S rRNA gene abundances across a given RBC flowpath is readily apparent (Fig. 2B), which would be unlikely if these genes did not represent the same population. Moreover, DGGE and sequencing of both thaumarchaeal amoA and 16S rRNA genes (Figs 5 and 6) suggest that a single detectable thaumarchaeal population inhabits this wastewater treatment system. It is also possible that higher thaumarchaeal 16S rRNA gene copy numbers result from archaeal populations that possess multiple 16S rRNA operons. However, given that all available AOA genomes contain only one 16S rRNA gene (Hallam et al., 2006; Walker et al., 2010; Blainey et al., 2011; Kim et al., 2011), it is more likely that low AOA amoA numbers result from poor primer matching with template DNA, consistent with previous findings (Konstantinidis et al., 2009). Indeed, although thaumarchaeal 16S rRNA genes were readily amplified from all samples, amoA genes could not be reliably amplified without use of a degenerate reverse primer and a decreased annealing temperature (data not shown). In this study, we considered archaeal amoA genes to be markers for AOA in an attempt to be conservative in our conclusions. However, based on thaumarchaeal 16S rRNA gene abundances, we have likely underestimated the abundance of AOA based on measured amoA gene copies, and therefore potential archaeal contributions to ammonia oxidation in this RBC treatment system.
In addition to qPCR data, this study quantified crenarchaeol as an independent metric of AOA abundance. Crenarchaeol is a glycerol dialkyl glycerol tetraether (GDGT) lipid that serves as a biomarker for AOA in ecological studies and has been detected in isolated and enriched AOA, including N. maritimus SCM1 (Schouten et al., 2008), Candidatus Nitrosocaldus yellowstonii (de la Torre et al., 2008), Candidatus Nitrososphaera gargensis (Pitcher et al., 2010) and Candidatus Nitrosoarchaeum limnia (Pitcher et al., 2011a). Intact polar lipids are found in cell membranes of living organisms, and are therefore considered to be biomarkers for viable cells. Upon cell death, polar head groups are readily hydrolysed, resulting in CLs, which are indicative of fossilized biomass. Lipid analyses are valuable and comparatively unbiased because of the lack of an amplification step. Indeed, Pitcher and colleagues (2011a) found strong congruence between crenarchaeol IPLs and the abundance and expression of thaumarchaeotal 16S rRNA in the Arabian Sea. In a representative full-length treatment train (June, NE train), both CL and IPL crenarchaeol abundances increased across the RBC flowpath (Fig. 4A). In addition, both core and IPL-derived crenarchaeol was higher in RBC 8 than RBC 1 for all treatment trains sampled (Fig. 4B). These data corroborate quantitative PCR results, and the high proportion of IPL-derived crenarchaeol suggests that the Thaumarchaea detected in this system represent viable cells, which are most likely contributing to ammonia oxidation. A few seasonal discrepancies were observed between lipid and genetic data. For example, crenarchaeol abundances were highest in June biofilm samples, whereas thaumarchaeal gene abundances were highest in September biofilm samples. However, direct comparison of genetic and lipid data can be problematic because measured gene abundances are relative values (i.e. copies per nanogram of genomic DNA), whereas lipid measurements represent absolute abundances (i.e. microgram of crenarchaeol per gram of biofilm). Taken together, the same overall trend is supported by both lipid and genetic data.
Although ammonium concentration correlated with AOA abundances, it had no observable effects on AOA diversity or community composition within the ammonia concentrations measured here (∼30–400 µg l−1). Thaumarchaeal amoA and 16S rRNA profiles contained one dominant band regardless of RBC stage, treatment train or season. Phylogenetic analyses of thaumarchaeal amoA and 16S rRNA genes retrieved from this RBC system demonstrated that archaeal sequences clustered with environmental sequences derived from other relatively high-ammonia environments, and share low homology with sequences from enriched or isolated AOA (Fig. 6). This finding may imply that lineages of AOA exist that are adapted to environments with varying nutrient status.
A variety of laboratory studies have identified ammonia as a key environmental parameter for determining relative abundances or activity of AOA and AOB, particularly in soil environments (Di et al., 2010; Taylor et al., 2010; Verhamme et al., 2011). In addition, laboratory incubations of soils have indicated that AOB are metabolically dominant following ammonium amendment (Jia and Conrad, 2009). The present study has taken advantage of an ammonia gradient created by existing wastewater treatment infrastructure and is unique because it provides a practical example of a phenomenon previously only identified through laboratory experiments.
To our knowledge, four previous studies have documented a relationship between ammonium availability and relative abundances of AOA and AOB in freshwater environments. Herrmann and colleagues (2011) demonstrated an increased ratio of AOB : AOA in simulated creek ecosystems amended with ammonia. In addition, Limpiyakorn and colleagues (2010) suggested ammonia as a potential variable in determining the relative abundances of AOA and AOB in municipal WWTPs, but this study produced no statistically significant correlations. A survey of Bangkok WWTPs reported a negative correlation between AOA amoA gene abundance in activated sludge and effluent ammonium concentrations (Kayee et al., 2011), but a similar correlation was not found with influent ammonium concentrations. A correlation was also observed between AOA : AOB ratios and ammonia concentration in freshwater aquarium biofilters (Sauder et al., 2011).
A recent study of municipal and industrial WWTPs found that AOA genes could rarely be detected in municipal WWTPs, but that in certain industrial WWTPs, thaumarchaeal amoA genes outnumbered bacterial amoA genes by up to four orders of magnitude (Mussmann et al., 2011). In one of these industrial plants, it was shown that the present thaumarchaea are not obligate chemolithoautotrophs, but instead may be using organic carbon for mixotrophic or heterotrophic metabolism. Although all currently cultured or enriched amoA-encoding thaumarchaeota oxidize ammonia (Könneke et al., 2005; Hatzenpichler et al., 2008; de la Torre et al., 2008; Tourna et al., 2011), this study by Mussmann and colleagues (2011) calls into question whether all Thaumarchaeota possessing amoA genes mediate ammonia oxidation. Therefore, we acknowledge the possibility that the Thaumarchaeota detected in the Guelph RBCs may be obtaining energy from organic carbon instead of, or in addition to, ammonia. Indeed, the 16S rRNA gene sequence retrieved from these RBCs clustered with clone sequences derived from industrial WWTPs analysed by Mussmann and colleagues (Fig. 6), although these sequences did not originate from the WWTP on which the majority of their study was based. Further activity studies and cultivation attempts will be necessary to confirm the role of Thaumarchaeota in tertiary wastewater treatment. Nonetheless, the Thaumarchaeota identified in this study demonstrate an adaptation to low-ammonia conditions, whether or not they are strict chemolithoautotrophs.
Because the discovery of AOA is a recent phenomenon, most existing nitrification infrastructure has been designed on the premise that it will host populations of AOB. However, this study suggests that AOA may play a role in ammonia oxidation in low-ammonia biofiltration systems, such as nitrifying RBCs, aquaculture, aquarium biofilters and drinking water treatment. These systems rely on nitrification for ammonia removal, but low ammonia concentrations may preclude AOB from obtaining sufficient energy for survival and growth. Existing studies support the hypothesis that AOA dominate low-ammonia engineered systems such as groundwater treatment and distribution systems (van der Wielen et al., 2009) and granular activated carbon of drinking water treatment plants (Kasuga et al., 2010). This may be important when designing biofiltration systems, because AOB and AOA likely vary in a variety of additional ecological adaptations. For example, previous studies have suggested that AOA thrive in low-oxygen conditions (Park et al., 2006; 2010). Therefore, the aggressive aeration typically provided in engineered biofilters (that would be beneficial for AOB) may actually interfere with the ability of AOA to oxidize ammonia as efficiently as possible.
The results of this study provide evidence for a low-ammonia niche of AOA within the RBC system in Guelph, Ontario. Future research will determine the extent to which ammonia concentration affects AOA : AOB ratios in additional WWTPs and biofiltration systems associated within additional engineered environments. This study also provides a foundation for future activity, cultivation and genomic analyses for characterizing nitrogen biogeochemistry within engineered freshwater systems.
Experimental procedures
Guelph WWTP design, sample collection and water chemistry analyses
The RBCs in the Guelph WWTP were designed for nitrification and are utilized for tertiary treatment following activated sludge treatment in aeration basins, and prior to sand filtration and chlorination (Fig. 1C). The Guelph WWTP features a total of 32 RBC stages arranged in four treatment trains, with each train situated in a tank that is 39.5 m in length, 8.0 m in width, and has a water depth of 1.6 m. Each treatment train consists of eight individual RBC stages, which wastewater passes through serially. The total medium surface area per RBC is 13 750 m2, which results in a combined surface area of 440 000 m2. Each RBC is approximately 40% submerged in secondary effluent, and continuous rotation at a velocity of 0.8 to 1.3 r.p.m. is driven by air via centrifugal blowers. The average hydraulic detention time across an RBC treatment train is 53 min.
Samples were collected from the Guelph WWTP (Guelph, Ontario, Canada), which is a full-scale municipal WWTP that serves a population of ∼120 000 and treats an average wastewater volume of 42 216 m3 per day (based on data from 2010). Samples were collected in February, June and September 2010. February samples were collected from all stages of the NE treatment train (Fig. 1C). In both June and September, all stages of both the NE and SE trains were sampled. Biofilm and RBC-associated wastewater were collected for each RBC stage. Each RBC contains sampling windows, allowing biofilm to be sampled directly from the internal medium surface. Biofilm samples were collected with an ethanol-treated spatula, stored in sterile plastic tubes, and placed on dry ice immediately, where they remained until transfer to −80°C storage. Water samples from each RBC were collected in sterile plastic tubes and stored on ice until return to the laboratory.
Dissolved oxygen (dO2) and water temperature were measured in situ using an HQ30d digital probe (Hach Company, Loveland, CO, USA). The pH was measured for all water samples using a DELTA 320 pH meter (Mettler Toledo, Mississauga, ON, Canada) directly upon return to the laboratory and prior to freezing. All water samples were then stored at −80°C, except samples used for DOC measurements, which were filtered (0.22-µm syringe) and stored in the dark at 5°C prior to DOC measurements. Dissolved organic carbon was measured using a Dohrman DC-190 High-Temperature TOC Analyser (Rosemount Analytical, Santa Clara, CA, USA). Samples were acidified using 20% phosphoric acid and sparged to remove dissolved inorganic carbon prior to analysis. Nitrate (NO3--N) concentrations were measured by ion chromatography using a Dionex ICS-90 (Dionex, Sunnyvale, CA, USA). Nitrite (NO2--N) concentrations were measured by colorimetric analysis using a DU 500 UV/Visible spectrophotometer (Beckman Coulter, Brea, CA, USA). All nitrate, nitrite and DOC analyses were performed in the Environmental Geochemistry Laboratory, Department of Earth and Environmental Sciences, University of Waterloo. Ammonium (NH4+-N) concentrations were determined fluorometrically, as outlined previously (Holmes et al., 1999), using a TD 700 fluorometer (Turner Designs, Sunnyvale, CA, USA).
DNA extraction and quantification
Genomic DNA was extracted from biofilm samples using the PowerSoil DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA, USA) as outlined in the manufacturer's instructions. Genomic DNA extracts were visualized on a 1% agarose gel by standard gel electrophoresis and quantified spectrophotometrically using a NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA).
Quantitative PCR
Quantification of AOA and AOB amoA genes used primers CrenamoA23F and a degenerated version of CrenamoA616R (Nicol et al., 2008), and amoA-1F and amoA-2R (Rotthauwe et al., 1997) respectively. Thaumarchaeal and general bacterial 16S rRNA genes were quantified using primers 771F and 957R (Ochsenreiter et al., 2003) and 341F and 518R (Muyzer et al., 1993) respectively. All qPCR amplifications were conducted in duplicate on a CFX96 system (Bio-Rad, Hercules, CA, USA). Each reaction volume of 12.6 µl contained 2× iQ SYBR Green Supermix (Bio-Rad), 5 pmol of each primer, 5 µg of bovine serum albumin and 1–10 ng of genomic DNA as template. This template concentration represented a 10-fold dilution of the extracted genomic DNA, and qPCR for serial dilutions of genomic DNA indicated no inhibition at this dilution. For AOA amoA genes, the PCR conditions were 95°C for 3 min followed by 35 cycles of 95°C for 30 s, 53°C for 30 s and 72°C for 1 min, with a fluorescence reading following each elongation step. For AOB amoA genes, PCR conditions were the same, but with an annealing temperature of 58°C. For both archaeal and bacterial 16S rRNA genes, PCR conditions were the same but with an annealing temperature of 55°C and an elongation time of 30 s. Standard curves were constructed using 10-fold serial dilutions of template DNA of known concentration. For all genes, template DNA consisted of PCR amplicons generated from the same primer pair used for qPCR. For AOA and AOB amoA and thaumarchaeal 16S rRNA gene amplicons, the original template source was a freshwater aquarium biofilter (Sauder et al., 2011). For bacterial 16S rRNA genes, the original template source was Escherichia coli genomic DNA.
Polymerase chain reaction amplification efficiencies ranged from 80.2% to 98.1% and all R2 values were greater than 0.99. For all amplification reactions, melt curves were performed from 65°C to 95°C with an incremental increase in temperature of 0.5°C. Polymerase chain reaction specificity was verified for all reactions using melt peaks and standard 1% agarose gel electrophoresis.
Lipid analysis
Biofilm samples were freeze-dried and extracted (3×) using a modified Bligh and Dyer (1959) technique. A solvent mixture of methanol (MeOH) : dichloromethane (DCM) : K phosphate buffer at pH 7.4 (2:1:0.8, v/v/v) was added to the sample in a centrifuge tube and placed in an ultrasonic bath for 10 min. The extract was collected after centrifuging the sample at 2500 r.p.m. for 2 min. Dichloromethane and phosphate buffer were added to the combined extracts to a new volume ratio of 1:1:0.9 (v/v/v) to achieve phase separation. The organic DCM phase and aqueous MeOH/phosphate buffer phase were separated by centrifuging at 2500 r.p.m. for 2 min. The DCM phase, containing the lipids, was passed over extracted cotton to remove possible remaining particles and collected in a glass tube. The aqueous phase was subsequently rinsed twice with DCM, and all cleaned DCM phases were combined and dried under a N2 flow and stored at −20°C until analysis.
The extracts were separated into a CL and an IPL fraction over an activated silica column according to Oba and colleagues (2006) and Pitcher and colleagues (2009), except that hexane : ethyl acetate (1:1, v/v) was used to retrieve the CLs, and that MeOH was used to obtain the IPLs. A C46 internal GDGT standard (0.1 µg) was added to the CL fraction and an aliquot of the IPL fraction according to Huguet and colleagues (2006), after which the IPL aliquot was subjected to acid hydrolysis to cleave all ether-bound and most of the ester-bound head groups and release their CLs (IPL-derived GDGTs).
Subsequently, the CL and IPL-derived fractions were dissolved in hexane : isopropanol (99:1, v/v), filtered over a 0.45 µm PTFE filter, and concentrated to ∼3 mg ml−1 prior to analysis using HPLC/atmospheric pressure chemical ionization–MS on an Agilent 1100 series LC/MSD SL according to Schouten and colleagues (2007), with minor modifications. In short, component separation was achieved with an Alltech Prevail Cyano column (150 mm × 2.1 mm; 3 µm). The GDGTs were eluted isocratically with 90% A and 10% B for 5 min and then with a linear gradient to 16% B for 34 min, where A = hexane and B = hexane : isopropanol (9:1, v/v). The injection volume for all samples was 10 µl. Single ion monitoring of (M + H)+-ions was used to detect and quantify the GDGTs. Absolute quantification was performed as described by Huguet and colleagues (2006), in which a typical analytical standard deviation of 5% was reported.
DGGE and band sequencing
DGGE fingerprinting was performed for AOA and AOB amoA genes, as well as thaumarchaeal and general bacterial 16S rRNA genes. DGGE for AOA amoA genes was performed as described previously (Tourna et al., 2008) with minor modifications. The amoA genes were amplified using primers CrenamoA23f and a degenerated version of crenamoA616R, and template concentrations ranged from approximately 0.5 ng to 10 ng per reaction. Polymerase chain reaction conditions were as described previously, except with an annealing temperature of 53°C. Thaumarchaeal 16S rRNA genes were amplified using primers 771F and 957R-GC, with amplification and DGGE conditions as outlined previously (Tourna et al., 2008). AOB amoA genes were amplified in a nested PCR approach, using primers amoA-1F and amoA-2R, followed by amoA1F-GC and amoA-2R, as outlined by Chu and colleagues (2007). For general bacterial 16S rRNA genes, amplification was performed using primers 341F-GC and 518R, and DGGE was performed as outlined previously (Muyzer et al., 1993).
All gels were run at 60°C and 85 V for 900 min, except general bacterial 16S rRNA gels, which were run for 840 min. The DGGE system used was a DGGEK-2401 (C.B.S. Scientific Company, Del Mar, CA, USA) using previously described technical modifications (Green et al., 2010). Gels were stained with SYBR green (Invitrogen) for 1 h, then scanned using the Typhoon 9400 Variable Mode Imager (GE Healthcare, Piscataway, NJ, USA) or the PharosFX (Bio-Rad). From the original gel images for each gene fragment analysed, fingerprints were normalized and aligned with GelCompar II (Applied Maths, Austin, TX, USA).
For thaumarchaeal amoA and 16S rRNA genes, DGGE bands were excised, amplified (using the above primers and conditions), and sequenced. Amplified DGGE bands were run on a second gel to ensure that each sequenced band corresponded to the original fingerprint. Because DGGE band sequences arising from thaumarchaeal 16S rRNA genes were short (i.e. < 200 bp), a longer thaumarchaeal 16S rRNA gene sequence was obtained with the archaeal primers 21F (DeLong, 1992) and 957R for the purpose of phylogenetic analysis. The longer sequence encompassed the 158 bp region of the corresponding DGGE band sequence; the sequences were identical across this span, indicating that they represented the same AOA population. These DNA sequences have been deposited in GenBank under accession numbers JN695686 and JN695687 for amoA and 16S rRNA genes respectively.
Sequences for thaumarchaeal amoA and 16S rRNA genes were compared with reference sequences (obtained from GenBank) of enriched or isolated AOA representatives as well as environmental representatives. Sequences were aligned using MUSCLE (Edgar, 2004), and the resulting alignments were cropped so that all sequences spanned the same 483-bp and 762-bp regions for amoA and 16S rRNA genes respectively. Evolutionary histories were inferred by using the Maximum Likelihood method based on the Tamura–Nei model of sequence evolution (Tamura and Nei, 1993). The trees shown were those with the highest log likelihood. Bootstrap testing was conducted with 500 replicates. All alignments and phylogenetic analyses were conducted in MEGA5 (Tamura et al., 2011).
Acknowledgments
We thank Nancy Evans and Tim Robertson (Guelph WWTP) and José Bicudo (Region of Waterloo) for their expertise and enthusiastic support of our study. Thank you to Katja Engel and Puntipar Sonthiphand for assistance with sampling and helpful suggestions. We acknowledge Richard Elgood, Andre Masella and Jort Ossebaar (NIOZ) for technical assistance and Cornelia Wuchter for suggestions that have improved the quality of this research. This research was funded by an Alexander Graham Bell Canada Graduate Scholarship to L. A. S. and a Discovery Grant to J. D. N., both provided by the National Sciences and Engineering Research Council of Canada (NSERC).
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Bacterial amoA (A) and general bacterial 16S rRNA (B) gene copies in biofilm samples across RBC flowpaths. Error bars represent standard deviations based on technical duplicates; error bars that are not seen are contained within the symbols.
Fig. S2. DGGE fingerprints for bacterial and thaumarchaeal amoA and 16S rRNA genes across eight serial RBC stages for all sampling seasons and RBC treatment trains.
Table S1. Water chemistry data for RBC-associated wastewater.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Beman JM, Popp B, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:429–441. doi: 10.1038/ismej.2007.118. [DOI] [PubMed] [Google Scholar]
- Beman JM, Francis CA. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl Environ Microbiol. 2006;72:7767–7777. doi: 10.1128/AEM.00946-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sørensen KB, Anderson R, et al. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA. 2006;103:3846–3851. doi: 10.1073/pnas.0600035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- Chu H, Fujii T, Morimoto S, Lin X, Yagi K, Hu J, Zhang J. Community structure of ammonia-oxidizing bacteria under long-term application of mineral fertilizer and organic manure in a sandy loam soil. Appl Environ Microbiol. 2007;73:485–491. doi: 10.1128/AEM.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Corte D, Yokokawa T, Varela MM, Agogué H, Herndl GJ. Spatial distribution of Bacteria and Archaea and amoA gene copy numbers throughout the water column of the Eastern Mediterranean Sea. ISME J. 2008;3:147–158. doi: 10.1038/ismej.2008.94. [DOI] [PubMed] [Google Scholar]
- DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di HJ, Cameron KC, Shen J-P, Winefield CS, O'Callaghan M, Bowatte S, He J-Z. Ammonia-oxidizing bacteria and archaea grow under contrasting soil nitrogen conditions. FEMS Microbiol Ecol. 2010;72:386–394. doi: 10.1111/j.1574-6941.2010.00861.x. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli K, Bosshard F, Werlen C, Lais P, Siegrist H, Zehnder AJ, van der Meer JR. Microbial composition and structure of a rotating biological contactor biofilm treating ammonium-rich wastewater without organic carbon. Microb Ecol. 2003;45:419–432. doi: 10.1007/s00248-002-2037-5. [DOI] [PubMed] [Google Scholar]
- Erguder T, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- Francis CA, Roberts K, Beman J, Santoro A, Oakley B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SJ, Leigh MB, Neufeld JD. Denaturing gradient gel electrophoresis (DGGE) for microbial community analysis. In: Timmis KN, editor. Microbiology of Hydrocarbon and Lipid Microbiology. Berlin, Germany: Springer-Verlag Berlin Heidelberg; 2010. pp. 4137–4158. [Google Scholar]
- Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann M, Scheibe A, Avrahami S, Kusel K. Ammonium availability affects the ratio of ammonia-oxidizing bacteria to ammonia-oxidizing archaea in simulated creek ecosystems. Appl Environ Microbiol. 2011;77:1896–1899. doi: 10.1128/AEM.02879-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RM, Aminot A, Kérouel R, Hooker BA, Peterson BJ. A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can J Fish Aquat Sci. 1999;56:1801–1808. [Google Scholar]
- Hou Y, Zhang H, Miranda L, Lin S. Serious overestimation in quantitative PCR by circular (supercoiled) plasmid standard: microalgal pcna as the model gene. PLoS ONE. 2010;5:e9545. doi: 10.1371/journal.pone.0009545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet C, Hopmans EC, Febo-Ayala W, Thompson DH, Sinninghe Damsté JS, Schouten S. An improved method to determine the absolute abundance of glycerol dibiphytanyl glycerol tetraether lipids. Org Geochem. 2006;37:1036–1041. [Google Scholar]
- Jang A, Okabe S, Watanabe Y, Kim IS, Bishop PL. Measurement of growth rate of ammonia oxidizing bacteria in partially submerged rotating biological contactor by fluorescent in situ hybridization (FISH) J Environ Eng Sci. 2005;4:413–420. [Google Scholar]
- Jia Z, Conrad R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- Jin T, Zhang T, Yan Q. Characterization and quantification of ammonia-oxidizing archaea (AOA) and bacteria (AOB) in a nitrogen-removing reactor using T-RFLP and qPCR. Appl Microbiol Biotechnol. 2010;87:1167–1176. doi: 10.1007/s00253-010-2595-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga I, Nakagaki H, Kurisu F, Furumai H. Predominance of ammonia-oxidizing archaea on granular activated carbon used in a full-scale advanced drinking water treatment plant. Water Res. 2010;44:5039–5049. doi: 10.1016/j.watres.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Kayee P, Sonthiphand P, Rongsayamanont C, Limpiyakorn T. Archaeal amoA genes outnumber bacterial amoA genes in municipal wastewater treatment plants in Bangkok. Microb Ecol. 2011;62:776–788. doi: 10.1007/s00248-011-9893-9. [DOI] [PubMed] [Google Scholar]
- Kim BK, Jung MY, Yu DS, Park SJ, Oh TK, Rhee SK, Kim JF. Genome sequence of an ammonia-oxidizing soil archaeon, ‘Candidatus Nitrosoarchaeum koreensis’ MY1. J Bacteriol. 2011;193:5539–5540. doi: 10.1128/JB.05717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol G, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- Limpiyakorn T, Sonthiphand P, Rongsayamanont C, Polprasert C. Abundance of amoA genes of ammonia-oxidizing archaea and bacteria in activated sludge of full-scale wastewater treatment plants. Bioresour Technol. 2010;102:694–701. doi: 10.1016/j.biortech.2010.11.085. [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- Mosier AC, Francis CA. Relative abundance and diversity of ammonia oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ Microbiol. 2008;10:3002–3016. doi: 10.1111/j.1462-2920.2008.01764.x. [DOI] [PubMed] [Google Scholar]
- Mussmann M, Brito I, Pitcher A, Sinninghe Damste JS, Hatzenpichler R, Richter A, et al. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc Natl Acad Sci USA. 2011;108:16771–16776. doi: 10.1073/pnas.1106427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- Oba M, Sakata S, Tsunogai U. Polar and neutral isopranyl glycerol ether lipids as biomarkers of archaea in near-surface sediments from the Nankai Trough. Org Geochem. 2006;37:1643–1654. [Google Scholar]
- Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ Microbiol. 2003;5:787–797. doi: 10.1046/j.1462-2920.2003.00476.x. [DOI] [PubMed] [Google Scholar]
- Park BJ, Park SJ, Yoon DN, Schouten S, Sinninghe Damsté JS, Rhee SK. Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl Environ Microbiol. 2010;76:7575–7587. doi: 10.1128/AEM.01478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-D, Wells GF, Bae H, Criddle CS, Francis CA. Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl Environ Microbiol. 2006;72:5643–5647. doi: 10.1128/AEM.00402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher A, Hopmans EC, Schouten S, Sinninghe Damsté JS. Separation of core and intact polar archaeal tetraether lipids using silica columns: insights into living and fossil biomass contributions. Org Geochem. 2009;40:12–19. [Google Scholar]
- Pitcher A, Rychlik N, Hopmans EC, Spieck E, Rijpstra WIC, Ossebaar J, et al. Crenarchaeol dominates the membrane lipids of Candidatus Nitrososphaera gargensis, a thermophilic Group I.1b Archaeon. ISME J. 2010;4:542–552. doi: 10.1038/ismej.2009.138. [DOI] [PubMed] [Google Scholar]
- Pitcher A, Hopmans EC, Mosier AC, Park SJ, Rhee SK, Francis CA, et al. Core and intact polar glycerol dibiphytanyl glycerol tetraether lipids of ammonia-oxidizing archaea enriched from marine and estuarine sediments. Appl Environ Microbiol. 2011a;77:3468–3477. doi: 10.1128/AEM.02758-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher A, Villanueva L, Hopmans EC, Schouten S, Reichart GJ, Sinninghe Damsté JS. Niche segregation of ammonia-oxidizing archaea and anammox bacteria in the Arabian Sea oxygen minimum zone. ISME J. 2011b;5:1896–1904. doi: 10.1038/ismej.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser JI, Nicol GW. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol. 2008;10:2931–2941. doi: 10.1111/j.1462-2920.2008.01775.x. [DOI] [PubMed] [Google Scholar]
- Pynaert K, Smets BF, Wyffels S, Beheydt D, Siciliano SD, Verstraete W. Characterization of an autotrophic nitrogen-removing biofilm from a highly loaded lab-scale rotating biological contactor. Appl Environ Microbiol. 2003;69:3626–3635. doi: 10.1128/AEM.69.6.3626-3635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotthauwe J, Witzel K, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauder LA, Engel K, Stearns JC, Masella AP, Pawliszyn R, Neufeld JD. Aquarium nitrification revisited: Thaumarchaeota are the dominant ammonia oxidizers in freshwater aquarium biofilters. PLoS ONE. 2011;6:e23281. doi: 10.1371/journal.pone.0023281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleper C. Ammonia oxidation: different niches for bacteria and archaea? ISME J. 2010;4:1092–1094. doi: 10.1038/ismej.2010.111. [DOI] [PubMed] [Google Scholar]
- Schouten S, Huguet C, Hopmans EC, Kienhuis MVM, Sinninghe Damsté JS. Analytical methodology for TEX86 paleothermometry by high-performance liquid chromatography/atmospheric pressure chemical ionization-mass spectrometry. Anal Chem. 2007;79:2940–2944. doi: 10.1021/ac062339v. [DOI] [PubMed] [Google Scholar]
- Schouten S, Hopmans EC, Baas M, Boumann H, Standfest S, Könneke M, et al. Intact membrane lipids of ‘Candidatus Nitrosopumilus maritimus’, a cultivated representative of the cosmopolitan mesophilic group I crenarchaeota. Appl Environ Microbiol. 2008;74:2433–2440. doi: 10.1128/AEM.01709-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Stehr G, Böttcher B, Dittberner P, Rath G, Koops H. The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol Ecol. 1995;17:177–186. [Google Scholar]
- Sturt HF, Summons RE, Smith K, Elvert M, Hinrichs KU. Intact polar membrane lipids in prokaryotes and sediments deciphered by high performance liquid chromatography/electrospray ionization multistage mass spectrometry – new biomarkers for biogeochemistry and microbial ecology. Rapid Commun Mass Spectrom. 2004;18:617–628. doi: 10.1002/rcm.1378. [DOI] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AE, Zeglin LH, Dooley S, Myrold DD, Bottomley PJ. Evidence for different contributions of Archaea and Bacteria to the ammonia-oxidizing potential of diverse Oregon soils. Appl Environ Microbiol. 2010;76:7691–7698. doi: 10.1128/AEM.01324-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- Tourna M, Freitag TE, Nicol GW, Prosser JI. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ Microbiol. 2008;10:1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x. [DOI] [PubMed] [Google Scholar]
- Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeisterc A, Urich T, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhamme DT, Prosser JI, Nicol GW. Ammonia concentration determines differential growth of ammonia-oxidising archaea and bacteria in soil microcosms. ISME J. 2011;5:1067–1071. doi: 10.1038/ismej.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells G, Park H, Yeung C, Eggleston B, Francis CA, Criddle C. Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ Microbiol. 2009;11:2310–2328. doi: 10.1111/j.1462-2920.2009.01958.x. [DOI] [PubMed] [Google Scholar]
- van der Wielen PWJ, Voost S, van der Kooij D. Ammonia-oxidizing Bacteria and Archaea in groundwater treatment and drinking water distribution systems. Appl Environ Microbiol. 2009;75:4687–4695. doi: 10.1128/AEM.00387-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Ye L, Tong AHY, Shao MF, Lok S. Ammonia-oxidizing archaea and ammonia-oxidizing bacteria in six full-scale wastewater treatment bioreactors. Appl Microbiol Biotechnol. 2011;91:1215–1225. doi: 10.1007/s00253-011-3408-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


