Abstract
Background
For a stem cell population to exist over an extended period, a balance must be maintained between self-renewing (proliferating) and differentiating daughter cells. Within the Caenorhabditis elegans germ line, this balance is controlled by a genetic regulatory pathway, which includes the canonical Notch signaling pathway.
Results
Genetic screens identified the gene teg-1 as being involved in regulating the proliferation vs. differentiation decision in the C. elegans germ line. Cloning of TEG-1 revealed that it is a homolog of mammalian CD2BP2, which has been implicated in a number of cellular processes, including in U4/U6.U5 tri-snRNP formation in the pre-mRNA splicing reaction. The position of teg-1 in the genetic pathway regulating the proliferation vs. differentiation decision, its single mutant phenotype, and its enrichment in nuclei, all suggest TEG-1 also functions as a splicing factor. TEG-1, as well as its human homolog, CD2BP2, directly bind to UAF-1 U2AF65, a component of the U2 auxiliary factor.
Conclusions
TEG-1 functions as a splicing factor and acts to regulate the proliferation vs. meiosis decision. The interaction of TEG-1 CD2BP2 with UAF-1 U2AF65, combined with its previously described function in U4/U6.U5 tri-snRNP, suggests that TEG-1 CD2BP2 functions in two distinct locations in the splicing cascade.
Keywords: CD2BP2, TEG-1, stem cell proliferation, pre-mRNA splicing, UAF-1, U2AF65
Introduction
Stem cells provide the material necessary for tissue generation during development, and tissue replacement and repair in adult animals. The capacity of stem cells to contribute to these processes over an extended time period is tied to their ability to produce both self-renewing and differentiating daughter cells. Self-renewing daughter cells retain the stem cell characteristics of the parental cell; therefore, a pool of stem cells is maintained over time. The differentiating daughter cells are used to make the desired tissue. Maintaining the balance between self-renewing and differentiating daughter cells is key to proper stem cell function since a disruption in this balance prevents long term tissue generation; too little self-renewal results in the eventual depletion of the stem cell population, while too little differentiation results in not enough cells being made for proper tissue generation. Understanding how this balance between self-renewal and differentiation is maintained is an essential step in understanding stem cell biology and function.
The germ line of the nematode Caenorhabditis elegans has emerged as a powerful model to study how a stem cell population maintains the balance between self-renewal and differentiation (Hansen and Schedl, 2006; Kimble and Crittenden, 2007). In the C. elegans hermaphrodite, the germ line is contained in two gonad arms that meet at a common uterus. Populations of mitotically dividing (proliferating) cells exist at the ends of the two gonad arms distal from the uterus (Figure 1A). In the adult, each population consists of ~200–250 cells (Lamont et al., 2004; Killian and Hubbard, 2005), with cells progressing through the cell cycle approximately every ~6.5–8 hours (Fox et al., 2011). At least some of these proliferating cells are considered stem cells. Cells are kept in the proliferative state due to their close proximity to the Distal Tip Cells (DTCs), with a single DTC capping the distal end of each gonad arm. The proliferation promoting influence of the DTC is primarily accomplished through the conserved GLP-1/Notch signaling pathway (Hansen and Schedl, 2006; Kimble and Crittenden, 2007; Racher and Hansen, 2010). The DSL (Delta/Serrate/LAG-2) LAG-2 and APX-1 ligands are expressed on the surface of the DTC (Henderson et al., 1994; Tax et al., 1994; Nadarajan et al., 2009). When they bind the GLP-1/Notch receptor (Yochem and Greenwald, 1989), which is expressed on the surface of the germ cells (Crittenden et al., 1994), the cleaved intracellular portion of GLP-1 (referred to as GLP-1(INTRA)) is thought to enter the nucleus and bind to the CSL (CBF1/Suppressor of Hairless/LAG-1) LAG-1 transcription factor (Christensen et al., 1996; Mumm and Kopan, 2000) and the SEL-8/LAG-3 co-activator (Doyle et al., 2000; Petcherski and Kimble, 2000). This complex then activates genes necessary for the proliferative fate. As cells move along the gonad arm, away from the DTC, Notch signaling levels are thought to decrease, allowing cells to enter meiotic prophase. The first obvious indication that cells have entered into meiotic prophase is when the DNA takes on a crescent moon shape as chromosomes pair and synapse (Francis et al., 1995a; Dernburg et al., 1998; MacQueen and Villeneuve, 2001). Cells continue to move proximally while transiting through the various stages of meiotic prophase, eventually forming fully differentiated gametes (sperm or oocytes depending on the age of the hermaphrodite).
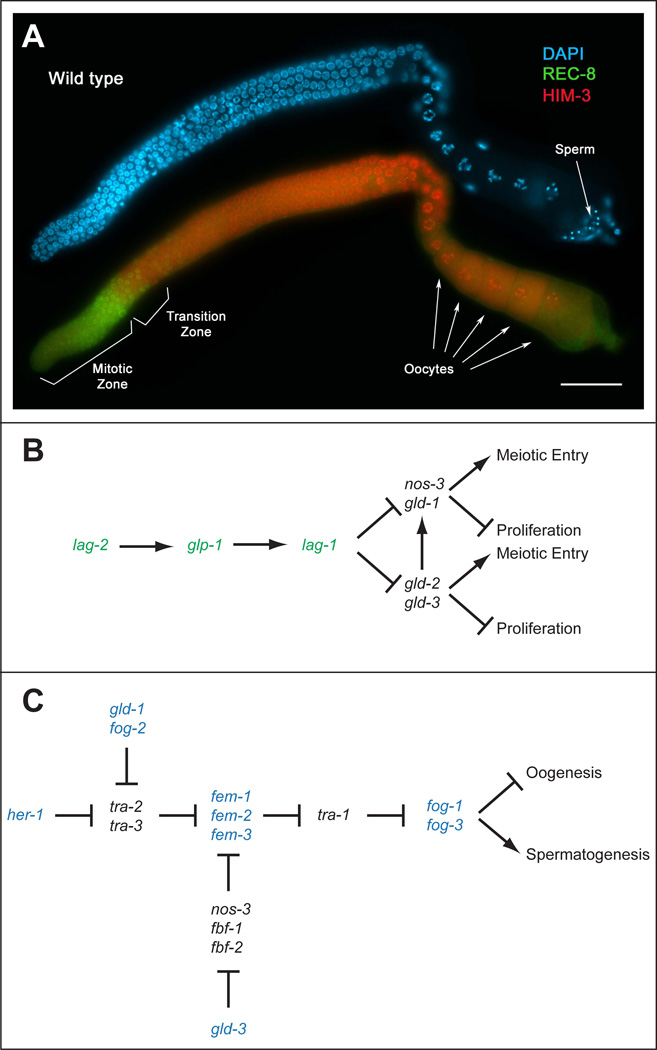
Figure 1.
The proliferation/differentiation balance and sex determination in the C. elegans germ line. (A) A wild-type adult (one day past the fourth larval stage) hermaphrodite dissected gonad arm stained with DAPI (blue) to visualize nuclear morphology, anti-REC-8 antibodies (green) to mark mitotically cycling cells, and anti-HIM-3 antibodies (red) to mark meiotic cells. Distal is to the left, with mitotic or proliferative cells existing in the distal end of the gonad arm. Cells move proximally, entering meiosis in the transition zone, and eventually forming first sperm, then oocytes. Scale bar =20 microns. (B) A simplified genetic pathway showing genes involved in regulating the proliferation vs. differentiation decision in the C. elegans germ line. Genes that promote proliferation are shown in green, while genes that inhibit proliferation and/or promote meiotic entry are shown in black. lag-2, glp-1 and lag-2, which encode components of the core Notch signaling pathway, through a number of intermediates, inhibit the activities of genes in two redundant downstream pathways, referred to as the gld-1 and gld-2 pathways. As cells move away from the distal end of the gonad, the gld-1 and gld-2 pathways become active, thereby promoting meiotic entry (differentiation) and/or inhibiting proliferation. Adapted from (Hansen and Schedl, 2006) (C) A simplified genetic pathway showing the genetic relationships between genes involved in regulating sex determination in the C. elegans germ line. Genes that promote the male fate are shown in blue, while genes that promote the female fate are shown in black. Hermaphrodites first produce sperm, and then a switch is made during development such that adult hermaphrodites produce oocytes. Adapted from (Ellis and Schedl, 2006).
The GLP-1/Notch signaling pathway promotes the proliferative fate, at least in part, by inhibiting the activities of two redundant meiotic promoting genetic pathways, referred to as the gld-1 and gld-2 pathways (Kadyk and Kimble, 1998) (Figure 1B). Within the gld-1 pathway are the gld-1 and nos-3 genes (Francis et al., 1995b; Hansen et al., 2004b); with gld-1 encoding a KH domain containing RNA binding translational regulator (Jones and Schedl, 1995), and nos-3 encoding a protein with similarity to Drosophila Nanos (Kraemer et al., 1999), also a translational regulator. Within the gld-2 pathway are the gld-2 and gld-3 genes, encoding the catalytic portion of a poly(A) polymerase (Kadyk and Kimble, 1998), and a homolog to Drosophila BicC, respectively (Eckmann et al., 2002). The spatial organization of the activity of the GLP-1/Notch signaling pathway relative to the activities of the gld-1 and gld-2 pathways is key to maintaining the balance between proliferation and differentiation. Loss or reduction of GLP-1/Notch signaling in the distal end results in ectopic activation of the gld-1 and gld-2 pathways in this region, causing cells to enter meiosis prematurely and a loss of the proliferative population (Austin and Kimble, 1987; Eckmann et al., 2004; Hansen et al., 2004b). Conversely, ectopic activation of GLP-1/Notch signaling beyond its normal region of activity, or a combined reduction in the activities of the redundant gld-1 and gld-2 pathways, results in excess proliferation (Berry et al., 1997; Kadyk and Kimble, 1998; Pepper et al., 2003; Hansen et al., 2004b). Indeed, in certain mutant gene and/or allele combinations, all cells within the gonad arm have the proliferative fate, resulting in a complete germline tumor (Berry et al., 1997; Hansen et al., 2004a). Other genes have been identified that function with the GLP-1/Notch signaling and gld-1/gld-2 pathways, or parallel to them, to regulate the balance between proliferation and differentiation. These genes function in many different cellular processes. For example, mutations that decrease the function of the proteasome enhance over-proliferation mutants, indicating a function to promote meiotic development (MacDonald et al., 2008). As another example, the cell cycle regulators Cyclin E (CYE-1) and CDK2 (CDK-2) promote the proliferative fate (Fox et al., 2011; Jeong et al., 2011). A number of splicing factors have also been identified as being involved in regulating the proliferation vs. differentiation decision (Belfiore et al., 2004; Mantina et al., 2009; Kerins et al., 2010; Zanetti et al., 2011). Genetically, these splicing factors appear to function largely in the gld-1 pathway, functioning downstream of GLP-1/Notch signaling (Figure 1B). A screen of splicing factors found that proteins functioning throughout the splicing cascade participate in the proliferation/differentiation decision, suggesting that an overall decrease in splicing or spliceosome activity contributes to over-proliferation (Kerins et al., 2010).
Pre-mRNA splicing involves the removal of introns from the pre-mRNA between the exons prior to nuclear export and translation. The splicing reaction is accomplished through the activity of the spliceosome, which is a large RNA-Protein complex that is assembled in a stepwise manner (Wahl et al., 2009; Valadkhan and Jaladat, 2010). Some of the earliest steps in spliceosome assembly include recognition of the 5’ and 3’ splice sites on the pre-mRNA by protein or RNA/protein complexes. The U1 snRNP particle, consisting of the U1 snRNA and a number of proteins, recognizes and binds to the 5’ splice site (Seraphin and Rosbash, 1989; Michaud and Reed, 1991). The U2 Auxiliary Factor (U2AF) recognizes and binds to the 3’ splice site (Wahl et al., 2009; Valadkhan and Jaladat, 2010). Human U2AF consists of two proteins, U2AF65 and U2AF35 (Zamore and Green, 1989), which in C. elegans are UAF-1 and UAF-2, respectively. U2AF65 binds to a polypyrimidine tract that is found at the 3’ end of introns, while U2AF35 binds to the 3’ splice site (Wahl et al., 2009; Valadkhan and Jaladat, 2010). Although in humans the polypyrimidine tract can vary somewhat in sequence and position relative to the 3’ splice site, in C. elegans the conserved UUUUCAG/R sequence is found immediately adjacent to the 3’ splice site (Blumenthal and Steward, 1997). Both UAF-1 and UAF-2 bind to this conserved sequence (Zorio and Blumenthal, 1999a). Binding of U2AF to the 3’ splice site helps in the recruitment of the U2 snRNP (Wahl et al., 2009; Valadkhan and Jaladat, 2010). In addition to recognition of the 5’ and 3’ splice sites by U1 and U2AF, respectively, SF1/BBP (SFA-1 in C. elegans (Mazroui et al., 1999)), binds to the branch point (Abovich and Rosbash, 1997; Berglund et al., 1997), which is just upstream from the polypyrimidine tract. Subsequent steps in spliceosomal assembly include replacement of SF1/BBP at the branch point by U2, recruitment of the U4/U6.U5 tri-snRNP, and disassociation of U1 and U2 from the spliceosome. This stepwise assembly contributes to the activated spliceosome (complex B*), which allows for the two transesterification reactions of intron removal to occur. The first transesterification reaction results in a free 5’exon and the 5’ end of the intron attached to the branch site in a lariat formation. The second transesterification reaction ligates the two exons and liberates the intron (Wahl et al., 2009; Valadkhan and Jaladat, 2010).
Here we demonstrate that reduction in teg-1 activity causes a disruption in the balance between proliferation and differentiation in the C. elegans germ line. In two different sensitized genetic backgrounds, teg-1 mutants were identified by their ability to enhance over-proliferation, resulting in a germ line tumor. We identified teg-1 as encoding a homolog of proteins from yeast to humans, including CD2BP2 and yeast Lin1p, which have been implicated in numerous cellular functions, including splicing. Within the splicing reaction CD2BP2 has been suggested to be involved in U4/U6.U5 tri-snRNP formation (Laggerbauer et al., 2005). We found that TEG-1 binds UAF-1, the worm homolog of the U2AF65 splicing factor, and that this interaction is conserved in humans. Therefore, TEG-1 CD2BP2 likely is involved in two distinct steps in the splicing reaction.
Results
teg-1 alleles identified in two over-proliferation enhancer screens
In an effort to better understand how the balance between proliferation and differentiation is controlled in the C. elegans germ line, we sought to identify genes involved in regulating this process. To identify these genes we performed two genetic screens for mutations that enhance germline over-proliferation. In the first screen, we mutagenized animals that contained the glp-1(oz112oz120) weak gain-of-function allele (Berry et al., 1997). These animals are relatively wild-type with respect to germline proliferation at 15°C, with only 0.05% of glp-1(oz112oz120) gonad arms containing an over-proliferative germ line (4/8000 gonad arms) (Berry, 1998). We screened for recessive enhancers of the over-proliferative phenotype by cloning F1 progeny of mutagenized animals and screening for those that showed ~25% of progeny (F2) with tumorous germ lines. From screening 8200 haploid genomes, we identified 12 enhancing mutations, with the strongest seven corresponding to four complementation groups. We have named these teg-1(oz189), teg-2(oz192, oz194, oz216, oz218), teg-3(oz190) and teg-4(oz210) (tumourous enhancer of glp-1(oz112oz120)). We have previously reported the cloning of teg-4, which encodes a homolog of the human SAP130 splicing factor (a subunit of the SF3b splicing complex) (Mantina et al., 2009). teg-2 and teg-3 are in various stages of characterization, mapping and cloning, which we will discuss elsewhere. Here we report the characterization and cloning of teg-1.
In the second screen, we identified mutations that have a synthetic over-proliferation phenotype with the gld-2(q497) allele. Since gld-2 pathway genes function redundantly with gld-1 pathway genes in regulating the balance between proliferation and differentiation (Kadyk and Kimble, 1998), and since the activities of both the gld-1 and gld-2 pathways must be reduced in order for a germline tumour to be formed, this screen was specifically designed to identify genes that function in the gld-1 genetic pathway, although mutations in genes that function upstream of Notch signaling, or in a parallel pathway, may also cause over-proliferation in a gld-2(q497) background. Since gld-2(q497) homozygous animals are sterile due to defects in gametogenesis (Kadyk and Kimble, 1998), we mutagenized gld-2(q497) homozygotes that were fertile because they carried the gaDp1 free duplication, which contains a wild-type copy of gld-2. The progeny (F2) of cloned F1 animals were screened for those with germline over-proliferation. These tumorous animals presumably lost the gaDp1 free duplication carrying the wild-type copy of gld-2, and were homozygous for the newly generated mutation. Since these animals were sterile, we recovered the induced mutation from siblings that carried gaDp1[gld-2(+)], and were either homozygous or heterozygous for the new mutation. We called genes identified in this screen syt (synthetic tumorous). We previously reported two genes identified in this screen; syt-1, which is allelic to nos-3 (Hansen et al., 2004b), and pas-5, which encodes a subunit of the proteasome (MacDonald et al., 2008). From this screen we also isolated the teg-1(oz230) allele; therefore, we isolated two alleles of teg-1, each from independent screens with different genetic backgrounds.
Analysis of teg-1 over-proliferation
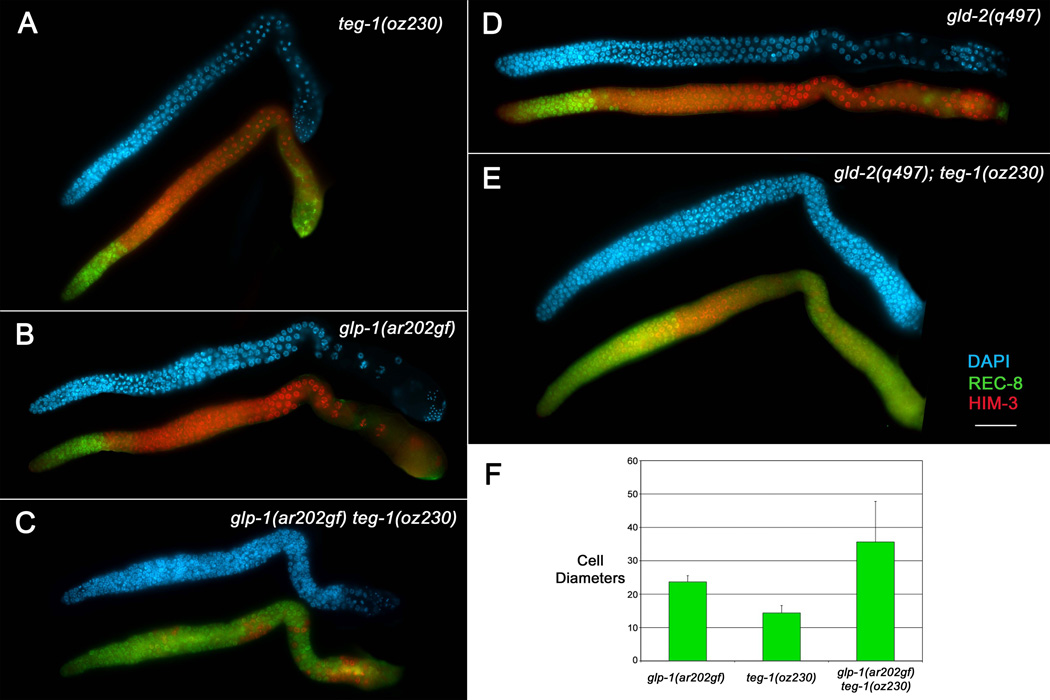
Since teg-1 mutant alleles were isolated from two different screens, we know that teg-1 mutants can cause/enhance over-proliferation in different sensitized genetic backgrounds. To more closely examine the nature of these over-proliferative tumors, we stained dissected gonads with markers for mitotic and meiotic cells; anti-REC-8 antibodies mark mitotic zone cells (including those in pre-meiotic S-phase) and anti-HIM-3 antibodies mark meiotic prophase cells (Zetka et al., 1999; Pasierbek et al., 2001; Hansen et al., 2004a). In performing the characterization we used the glp-1(ar202gf) allele for the sensitized background to be consistent with the analyses of other over-proliferation enhancing mutations (Hansen et al., 2004a; Hansen et al., 2004b; MacDonald et al., 2008; Mantina et al., 2009). We found that glp-1(ar202gf) teg-1(oz230) germ lines were over-proliferative, with excess REC-8(+) HIM-3(−) cells as compared to wild type or either single mutant (Figure 2). However, the excess proliferation was primarily found in the distal and proximal regions of the gonad, with meiotic cells found in between. The average size of the distal mitotic zone in glp-1(ar202gf) teg-1(oz230) double mutant animals was 35 cell diameters from the distal end, while the average sizes for the teg-1(oz230) and glp-1(ar202gf) single mutants were 14 and 22 cell diameters, respectively (Figure 2C, F). Therefore, the double mutant displays a late-onset-tumorous phenotype in which the size of the distal mitotic zone is larger than wild type, which has a mitotic zone ~20 cell diameters in length (Crittenden et al., 1994; Hansen et al., 2004a).
Figure 2.
teg-1(oz230) enhances the over-proliferative phenotype in different genetic backgrounds. Dissected gonad arms from adult hermaphrodite animals one day past the fourth larval stage. Gonad arms were stained with DAPI (blue) to show nuclear morphology, anti-REC-8 antibodies (green) to mark mitotic cells, and anti-HIM-3 antibodies (red) to mark meiotic cells. Both teg-1(oz230) (A) and glp-1(ar202gf) (B) animals, grown at 15°C, show proliferation restricted to the distal part of the gonad arm (although some anti-REC-8 cross-reactivity with sperm is observed in the proximal ends of the gonad arms); however, mitotic cells are found throughout the gonad arms of glp-1(ar202gf) teg-1(oz230) (C) animals (100% of gonad arms show this over-proliferation phenotype, n>20), although clusters of cells that have entered into meiosis are observed (red). (D–E) teg-1(oz230) also enhances the over-proliferative phenotype of gld-2(q497) single mutants (D), with gld-2(q497); teg-1(oz230) double mutants displaying extensive proliferation in both the distal and proximal regions of the gonad arms (E) (100% of gonad arms show this over-proliferation phenotype, n>20). All strains were also marked with unc-32(e189). Scale bar =20 microns. (F) The overall size of the of the distal mitotic zone is statistically larger in glp-1(ar202gf) teg-1(oz230) animals than in either single mutant, as measured by the number of cell diameters from the distal end of the gonad arm that are mitotic [anti-REC-8(+) (green) and distal to any meiotic nuclei -anti-HIM-3(+); (red)] (P < 9.7×10-5 t test. Error bars =1 SD. n= 20 gonad arms).
As has been observed with other mutations that are synthetic tumorous with gld-2(q497), gld-2(q497); teg-1(oz230) animals, although over-proliferative, have some cells that enter meiotic prophase (Figure 2E). The proportion of cells that appear meiotic differ from animal to animal; however, all gonad arms have some cells that have entered into meiosis (anti-HIM-3(+)). Therefore, the combined removal of gld-2 and teg-1 function does not completely shift the balance towards proliferation.
teg-1 encodes a homolog of human CD2BP2
To determine how teg-1 may be functioning at the molecular level, we mapped and cloned the gene. teg-1(oz230) was localized to a 0.4 cM region between spe-16 and dpy-18 on linkage group III by standard three-factor mapping. Single nucleotide polymorphism and deficiency mapping further narrowed the critical region to ~25 kb containing three genes. Sequencing revealed that one these genes, Y47D3A.27, contained premature stop codons associated with each of the two teg-1 alleles (Figure 3). An integrated transgene containing the Y47D3A.27 gene rescues the mutant phenotype of teg-1(oz230) (see below). Additionally, antibodies raised against the predicted Y47D3A.27 protein, which detects a protein of the correct size in wild-type extracts, fails to detect a protein in teg-1(oz230) mutant extracts (see below). Together, these data support teg-1 as being Y47D3A.27.
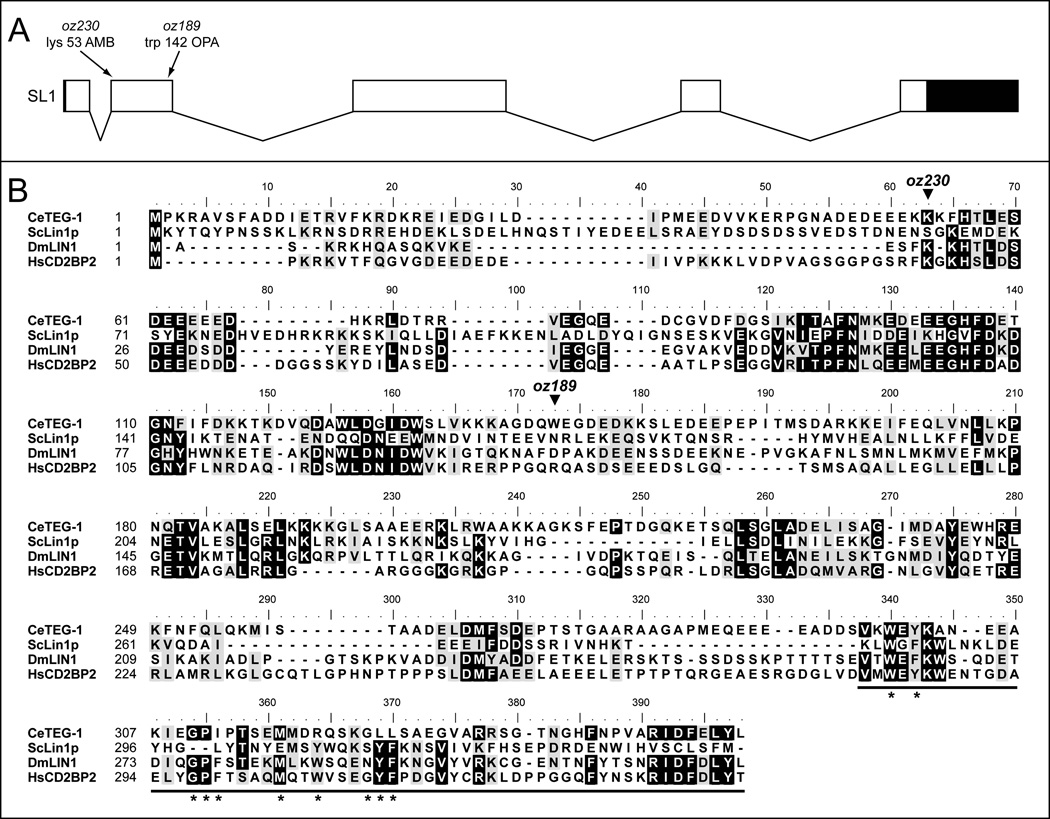
Figure 3.
TEG-1 is homologous to proteins from yeast to humans. (A) Sequencing of the yk82g9 cDNA, analysis of contig MM454_contig00336, as well as sequencing reverse transcribed PCR amplification products, revealed a gene model for teg-1 that includes five exons. Boxes represent exons, while joining lines represent introns. Blackened portions of the exons represent non-coding regions of the exons (5; and 3’ UTRs). From the A in the initiator ATG, the first exon includes nucleotides from position 1–138, exon two 239–544, exon three 1948–2220, exon four 3106–3315 (the gene model contained in wormbase places exon four at 3106–3323), exon five 4218–4352 (NOTE- only includes coding portions of exons, including taa stop codon). 5’RACE and 3’RACE revealed a 7bp 5’UTR and a 479bp 3’UTR, and also revealed that the teg-1 transcript is SL1 trans-spliced. The positions of the mutations associated with the two alleles of teg-1 are shown, with the oz230 mutation changing AAG to UAG (amber) at codon 53 (position 257 on genomic DNA from ATG). The oz189 mutation changes UGG to UGA (opal) at codon 142 (position 526 on genomic DNA from ATG). (B) An alignment of amino acid sequences between the TEG-1 predicted protein and homologs from Saccharomyces cerevisiae, Drosophila melanogaster and humans. Alignment was performed using ClustalW, with similar amino acids shown with black text on a grey background, and identical amino acids shown with white text on a black background. Amino acids were considered similar or identical if three or four of the four proteins contained a similar or identical amino acid at a given position. The thick black underline at the end of the proteins represents the regions deleted in the ΔGYF constructs. Asterisks are below each of the amino acids in CD2BP2 that are part of the GYF signature (W-X-Y-X6–11-G-P-F-X4-M-X2-W-X3-G-Y-F) (Kofler and Freund, 2006) The predicted locations of the premature stop codons introduced by the oz189 and oz230 alleles are shown. TEG-1 is 14.8%, 30.9% and 30.7% identical to ScLin1p, DmLIN1 and HsCD2BP2, respectively; and 25.6%, 42.2%, and 41.7% similar to these same proteins. Genbank accession numbers: ScLin1p (NP_012026.1), DmLin1 (Q9VKV5) and HsCD2BP2 (NP_006101.1).
Sequencing of a cDNA corresponding to teg-1, yk82g9, as well as sequencing RT-PCR generated cDNA using primers that are predicted to amplify the entire coding region, revealed a gene model differing slightly from the predicted gene model for Y47D3A.27 (http://ws220.wormbase.org/). Sequencing of these cDNAs suggest a gene model that contains five exons and is predicted to produce a protein of 353 amino acids (as compared to the predicted gene model that includes six exons and a 370 amino acid protein; Figure 3). The oz230 and oz189 alleles contain premature stop codons at amino acid positions 53 and 142, respectively. Therefore, oz189 and oz230 are likely strong loss-of-function or null alleles.
BLAST alignment of the predicted TEG-1 protein identifies homologous proteins from a variety of species, including CD2BP2 from humans, DmLIN1 from Drosophila melanogaster and Lin1p from Saccharomyces cerevisiae (Figure 3). Although conserved regions are found over the entire lengths of the proteins, the level of similarity is not overly high; TEG-1 is 42% similar and 31% identical to human CD2BP2, 42% similar and 31% identical to Drosophila DmLIN1, and 26% similar and 15% identical to yeast Lin1p. CD2BP2 homologs have been suggested to function in a number of processes, including pre-mRNA splicing, DNA replication, chromosome segregation and the mammalian immune response (Nishizawa et al., 1998; Freund et al., 1999). CD2BP2 homologs do not have any obvious motifs, other than a GYF motif in the carboxy end. The GYF domain was first identified in CD2BP2, and binds proline rich sequences (Nishizawa et al., 1998; Freund et al., 1999). Alignment of proteins from numerous species revealed a sequence signature for the domain; W-X-Y-X6–11-G-P-F-X4-M-X2-W-X3-G-Y-F, where X denotes any amino acid (Kofler and Freund, 2006). TEG-1 does contain some sequence similarity to the GYF signature in its carboxy end; however, it does not completely match the signature (Figure 3). Therefore, based solely on sequence similarity to the characterized GYF domain, it is not clear if TEG-1 contains this functional protein interaction domain in its carboxy terminus (see below).
TEG-1 controls germline development in a similar fashion to splicing factors
As mentioned, TEG-1 is homologous to CD2BP2 homologs, which have been suggested to have many functions. Perhaps the best-characterized function of these homologs is their role in pre-mRNA splicing, with CD2BP2 functioning in the formation of the U4/U6.U5 tri-snRNP (Laggerbauer et al., 2005). Interestingly, splicing factors have been shown to be involved in regulating the proliferation vs. differentiation decision in the C. elegans germ line (Puoti and Kimble, 1999; Puoti and Kimble, 2000; Belfiore et al., 2004; Konishi et al., 2008; Mantina et al., 2009; Kerins et al., 2010; Zanetti et al., 2011); therefore, teg-1’s involvement in this decision could be through a role in pre-mRNA splicing. However, the overall low level of similarity between TEG-1 and its homologs, as well as the other cellular functions for CD2BP2 homologs, prevent us from assuming that TEG-1 has a role in splicing. We reasoned that if TEG-1 is functioning as a splicing factor, and if it is a loss of this function in teg-1 mutants that contributes to the over-proliferation defect, teg-1 mutants should show similar phenotypes and epistatic relationships as other splicing factor mutants. To test this we first made double mutants with teg-1(oz230) and mutants of the genes functioning in the gld-1 and gld-2 pathways regulating the proliferation vs. meiotic entry decision. As with other splicing factor mutants, teg-1(oz230) forms synthetic tumors with both of the gld-2 pathway genes (gld-2 and gld-3), but not with the gld-1 pathway genes (gld-1 and nos-3; Table 1), suggesting that teg-1 likely functions in the gld-1 pathway. Additionally, the gld-3(q730); teg-1(oz230) tumor is epistatic to glp-1(q175null) (Table 1), suggesting that teg-1 functions downstream of Notch signaling, which is also consistent with what has been found with other splicing factor mutants (Mantina et al., 2009; Kerins et al., 2010).
Table 1.
Epistasis analysis of teg-1 with genes regulating the proliferation vs. differentiation decision.
| Genotype | Proliferation vs. Differentiation Phenotype (20°C)1 |
|---|---|
| teg-1(oz189 and oz230) | Wild-type |
| gld-1(q485) | Wild-type2 |
| nos-3(oz231) | Wild-type3 |
| gld-2(q497) | Wild-type4 |
| gld-3(q730) | Wild-type5 |
| glp-1(q175) | Glp6 |
| gld-2(q497) gld-1(q485) | Tumorous7 |
| gld-1(q485); teg-1(oz230) | Wild-type8 |
| nos-3(oz231); teg-1(oz230) | Wild-type9 |
| gld-2(q497); teg-1(oz230) | Tumorous10 |
| gld-3(q730); teg-1(oz230) | Tumorous |
| gld-2(q497) gld-1(q485); glp-1(q175) | Tumorous11 |
| gld-2(q497); glp-1(q175) teg-1(oz230) | Glp12 |
| gld-3(q730) ; glp-1(q175) teg-1(oz230) | Tumorous13 |
100% of animals show the phenotype described, unless otherwise stated. For each genotype, a minimum of 30 gonad arms was analyzed.
(Francis et al., 1995a). gld-1(q485) XX animals have tumorous germ lines; however, this has to do with female germ cells entering into meiotic prophase, but failing to progress through meiosis. These cells then return to mitosis, forming a tumor. The initial entry into meiosis in these animals is very similar to wild-type; therefore, they do not have a defect in the balance between proliferation and meiotic entry.
(Austin and Kimble, 1987). Germ line proliferation abnormal. The proliferative population of cells is depleted, resulting in gonads containing only a limited number of differentiated cells. In glp-1 animals, only ~16 sperm are formed per gonad arm.
Vab progeny from gld-1(q485)/hT2; vab-7(e1562) teg-1(oz230) were examined by Nomarski optics and dissected gonads were analyzed using fluorescent microscopy. Both XX and XO animals were examined.
Unc progeny from nos-3(oz231); unc-32(e189) teg-1(oz230) were examined by Nomarski optics
Vab or Unc self or cross progeny from gld-2(q497)/hT2; vab-7(e1562) teg-1(oz189 or oz230)/hT2 or gld-2(q497)/hT2; unc-32(e189) teg-1(oz189 or oz230)/hT2 were examined by Nomarski optics. Also, dissected gonads were DAPI stained and analyzed using fluorescent microscopy.
Premature entry to meiosis phenotype similar to a glp-1(null) phenotype. Unc progeny from gld-2(q497)/hT2; unc-32(e189) glp-1(q175 and q172) teg-1(oz230)/hT2 were examined by Nomarski optics and dissected gonads were analyzed using fluorescent microscopy.
Unc progeny from gld-3(q730)/mIn1; unc-32(e189) glp-1(q175) teg-1(oz230)/hT2 were examined by Nomarski optics and dissected gonads were analyzed using fluorescent microscopy.
Additionally, we also observed a difference in epistasis with gld-2 and gld-3, suggesting that their functions are not equivalent in the gld-2 pathway. While gld-3(q730); teg-1(oz230); glp-1(q175) animals have a tumorous germ line, gld-2(q497); glp-1(q175) teg-1(oz230) animals are not tumorous, but rather display the under-proliferative Glp phenotype associated with glp-1(q175) (Table 1). Similar epistasis results were observed for the teg-4 and prp-17 splicing factors (Mantina et al., 2009; Kerins et al., 2010). Tumor formation in gld-3(q730); glp-1(q175); teg-1(oz230), with a complete absence of GLP-/Notch signaling, strongly supports the placement of teg-1 downstream of Notch signaling in the genetic pathway regulating the proliferation vs. differentiation decision. The absence of tumor formation in gld-2(q497); glp-1(q175) teg-1(oz230) animals could still be consistent with teg-1 functioning downstream of GLP-1/Notch signalling, if teg-1 functions redundantly with another gene/pathway in inhibiting proliferation; for example, if removing teg-1 function does not completely remove the activity of the gld-1 pathway. That gld-2(q497); glp-1(q175) teg-1(oz230) animals are Glp, while gld-3(q730); glp-1(q175) teg-1(oz230) animals are tumorous, suggests that removal of gld-3 reduces the activity of the gld-2 pathway to a greater extent than the removal of gld-2 activity, or that gld-3 has functions in addition to its role in the gld-2 pathway. In any event, the fact that a synthetic tumor is formed in gld-3(q730); teg-1(oz230) animals, even in the absence of GLP-1/Notch signaling, strongly supports the placement of teg-1 downstream of GLP-1/Notch signaling. Additionally, the fact that teg-1 shows the same complex epistatic relationships as the teg-4 and prp-17 splicing factors supports teg-1 functioning as a splicing factor.
Characterization of teg-1 single mutants
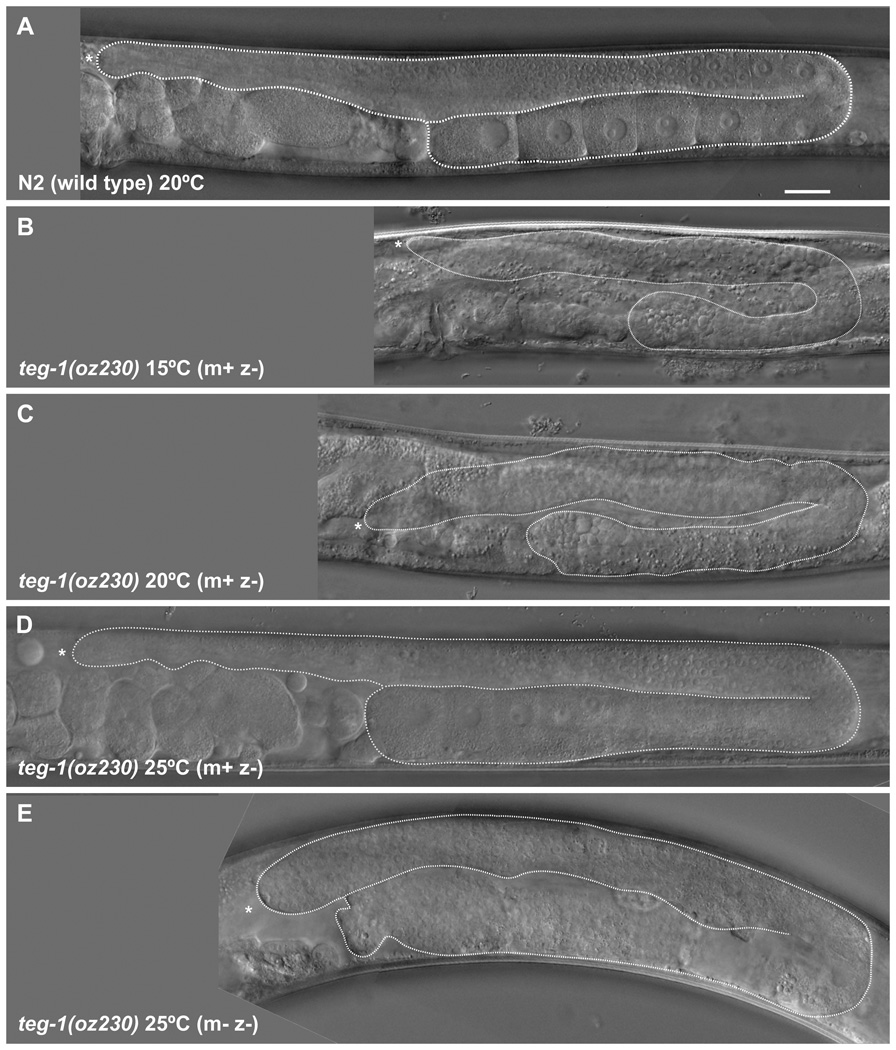
To further compare teg-1(0) phenotypes with those observed in other splicing factor mutants, we analyzed teg-1’s single mutant phenotypes. As with other splicing factor mutants (Belfiore et al., 2004; Mantina et al., 2009; Kerins et al., 2010; Zanetti et al., 2011), teg-1 has a role in germline sex determination; loss of teg-1 function results in a cold sensitive Mog (masculinization of the germ line) phenotype. At 15°C, teg-1 hermaphroditic animals fail to switch from the production of sperm to oocytes, resulting in over ~600 sperm made per gonad arm (632±156, n=5), as compared to ~150 in wild-type animals (Hirsh et al., 1976) (Figure 4). We also found that fog-1(q241), fog-3(q443) and fem-3(e1996) are all epistatic to teg-1(oz230) (Table 2), suggesting that teg-1 likely functions upstream of these genes. Epistasis of germline sex determination genes with other splicing factors, as well as somatic reporter assays with fem-3, suggest that the splicing factors could be functioning on the fem-3 gene, ultimately regulating its translation (Graham and Kimble, 1993; Graham et al., 1993; Ellis and Kimble, 1995; Gallegos et al., 1998; Kerins et al., 2010). A complexity to this is that we found that both fog-2(null) and tra-2(gain-of-function) are, at least partially, epistatic to teg-1(oz230) (Table 2). However, the epistasis with fog-2 is incomplete (88% of teg-1(oz230); fog-2(q71) animals are Fog (Feminization of the germ line), with the remaining animals showing some sperm production); therefore, teg-1 could still function with the other splicing factors in regulating fem-3, and the epistasis with fog-2(null) and tra-2(gf) could reflect a limited role for teg-1 in regulating fem-3. It must be noted that teg-1’s role in sex determination is not essential; homozygotes are not Mog at 25°C, even though the teg-1(oz189 & oz230) alleles are likely null (see below). Therefore, even at 15°C, knocking out teg-1 function likely does not completely block the sex determining signaling pathway at the position where teg-1 functions. An increase in a feminization signal upstream of teg-1, such as with fog-2(null) and tra-2(gf), could still result in sufficiently decreased activity of the fem genes so as to cause feminization.
Figure 4.
teg-1(oz230) single mutant animals display temperature specific germline phenotypes. Germline phenotypes of teg-1(oz230) homozygous animals were analyzed using Nomarski optics in adult animals one day past the fourth larval stage. Animals were grown at (B) 15°C, (C) 20°C and (D, E) 25°C. A wild-type animal (A) grown at 20°C is provided for comparison. For each animal, only one gonad arm is shown, with the distal arm on top, and the distal end to the left. The gonad arm is outlined with a dashed line, from the distal end to the spermatheca. Strains were grown as teg-1(oz230)/hT2 heterozygotes at the specified temperature for multiple generations prior to analyzing teg-1(oz230) homozygous animals for their germ line phenotype, except for teg-1(oz230 m-z-) animals (E), in which the mother of the analyzed animal was also homozygous for teg-1(oz230). (B) Gonads in animals grown at 15°C are somewhat smaller and exclusively produce sperm, even in older animals. (C) At 20°C, animals produce sperm and smaller misshapen oocytes; however, they are sterile. (D) At 25°C, animals produce sperm and oocytes, as well as viable progeny; however, the progeny (E) are sterile, with no obvious oocyte production, although a limited number of sperm are observed. For each temperature >100 animals were analyzed, with all animals grown at the same temperature exhibiting similar phenotypes. Actual genotype unc-32(e189) teg-1(oz230). Scale bar =20 microns.
Table 2.
Interaction of teg-1 with genes in the germ line sex determination pathway.
| Genotype | Germline Phenotype (15°C) |
|---|---|
| teg-1(oz189 and oz230) | Mog1 |
| fog-1(q241)2 | Fog3 |
| fog-2(q71)4 | Fog5 |
| fog-3(q443)6 | Fog |
| fem-3(e1996)7 | Fog |
| tra-2(e2020gf)8 | Fog |
| fog-1(q241); teg-1(oz230)9 | Fog |
| teg-1(oz230); fog-2(q71)10 | Fog |
| fog-3(q443); teg-1(oz230)11 | Fog |
| teg-1(oz189); fem-3(e1996)12 | Fog |
| tra-2(e2020gf); teg-1(oz230)13 | Fog |
Masculinization of the germ line
Feminization of the germ line
XX only- XO animals are wild type
All Vab Unc progeny of fog-1(q241) unc-13(e51)/ hT2; vab-7(e1562) teg-1(oz230)/hT2 were Fog.
fog-2(q71) rol-9(sc148)/ unc-51(e369) fog-2(q71) males mated with vab-7(e1562) teg-1(oz230)/hT2 hermaphrodites. L4 hermaphrodites cloned and put at 15°C. Of those that segregated both Rol and Vab animals, 21/24 Rol Vab progeny (vab-7(e1562) teg-1(oz230); fog-2(q71) rol-9(q71) contained small oocytes and no sperm, 3/24 contained sperm. 21/21 non-Rol non-Unc Vab animals (teg-1(oz230); fog-2(q71)/+) were Mog.
All Unc progeny of teg-1(oz230)/hT2; unc-13(e51) fog-3(oz230) were Fog.
vab-7(e1562) teg-1(oz189); unc-24(e138) fem-3(e1996) dpy-20(e1282) females were obtained from an unbalanced strain.
tra-2(e2020) males were mated with vab-7(e1562) teg-1(oz230)/hT2 purged hermaphrodites. Single male progeny were then mated with single female progeny. From matings that failed to segregate hT2, L4 (XX) Vab animals were cloned and scored the next day. 19/25 animals had small oocytes and no sperm. The remaining six animals were Mog (presumably tra-2(+)/tra-2(+)).
teg-1 single mutants also display other phenotypes. At 20°C, teg-1(0) animals are sterile, producing sperm and small, abnormally shaped oocytes (Figure 4), while at 25°C, they produce functional sperm and oocytes, and therefore are fertile, provided that their mother had one wild-type copy of teg-1 (in other words, m+z− animals are fertile). Homozygous progeny of homozygous mothers (m−z−), grown at 25°C, are sterile with no differentiated germ cells being formed (Figure 4). Therefore, in addition to the cold sensitive Mog phenotype, which is consistent with TEG-1 functioning as a splicing factor, TEG-1 also has functions in germline proliferation and oogenesis.
TEG-1 is enriched in the nucleus
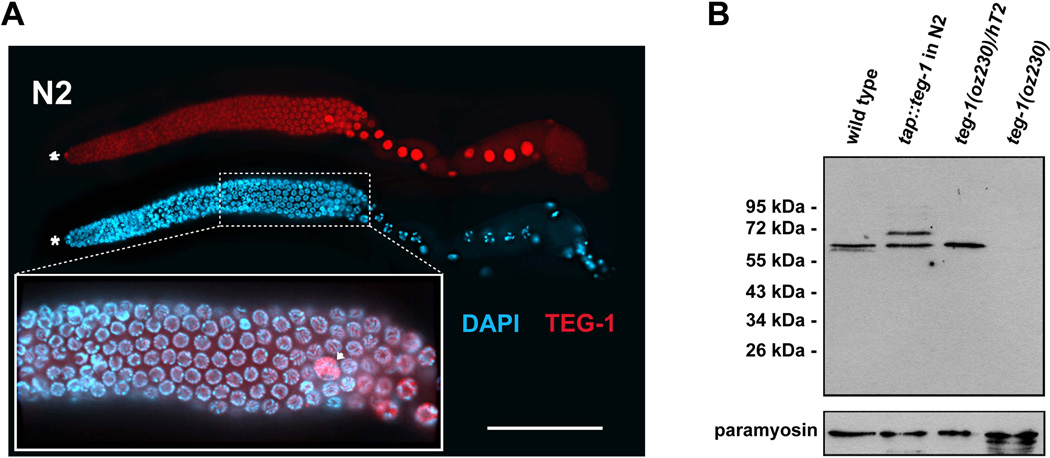
Splicing occurs in the nucleus of cells; therefore, most splicing factors are nuclear enriched. To determine if TEG-1 is also nuclear enriched, supporting a role for TEG-1 as a splicing factor, as well as to gain additional insight into TEG-1 function, we raised antibodies against TEG-1 (see Experimental Procedures). These antibodies are specific to TEG-1 as they detect an ~60 kD protein on western blots that is absent in teg-1(oz230) homozygotes (Figure 5). Staining revealed that TEG-1 is enriched in the nuclei of all germline and somatic cells of the gonad (Figure 5); however, staining above background was also detected in the cytoplasm of the gonad and intestine, suggesting that TEG-1 is also present in the cytoplasm (Figure 5). Within the nuclei, TEG-1 is found throughout the nucleoplasm, but appears to be absent in the regions containing DNA. TEG-1 levels are not uniform throughout the cells of the gonad. TEG-1 levels appear highest in nuclei of the oocytes, the somatic Distal Tip Cell (DTC), and sheath cells. TEG-1 enrichment in the nucleus is consistent with a role in splicing.
Figure 5.
TEG-1 is nuclear enriched in all gonadal cells. (A) A dissected gonad arm from a wild-type (N2) adult hermaphrodite, one day past the fourth larval stage grown at 20°C, stained with DAPI (blue) to visualize nuclear morphology, and anti-TEG-1 specific antibodies (red). The distal end of the gonad is to the left. Highest levels of TEG-1 enrichment are observed in nuclei of the distal tip cell (asterisk), sheath cells (arrow head) and oocytes (large nuclei in the proximal half of the gonad arm). The inset shows a blow up of a portion of the pachytene region of the gonad arm, with the DAPI and anti-TEG-1 channels merged. The regions of the nuclei occupied by DNA and TEG-1 appear to be mutually exclusive. Intensity comparisons of oocyte and intestine cytoplasm in wild-type and teg-1(oz230) animals demonstrates that some TEG-1 is present in the cytoplasm (oocyte intensity measurements- 1695±428 (n=10) in N2 vs. 456±258 (n=10) in teg-1(oz230)—Intestine intensity measurements- 1504±218 (n=15) in N2 vs. 596±233 (n=17) in teg-1(oz230)). Scale bar = 20 microns. (B) Western blot showing relative size of the TEG-1 protein, and the specificity of the anti-TEG-1 antibodies. A TAP-tag (HA-His8x-TEV-Myc) was fused to the N-terminus of teg-1 genomic DNA to generate an integrated transgenic line. Proteins at 65 kDa (endogenous TEG-1) and at 72 kDa (TAP-TEG-1) are detected in the extract of the transgenic strain. The comparable expression levels between these two proteins indicates that the TAP-TEG-1 is expressed at a level similar to endogenous TEG-1. Approximately 80 adult hermaphorodites from each C. elegans strain were loaded onto each lane of the SDS-PAGE gel. The wild-type and teg-1(oz230) strains are also marked with unc-32(e189). Paramyosin, detected with MH16 antibodies, was used as a loading control.
TEG-1 physically interacts with known splicing factor UAF-1
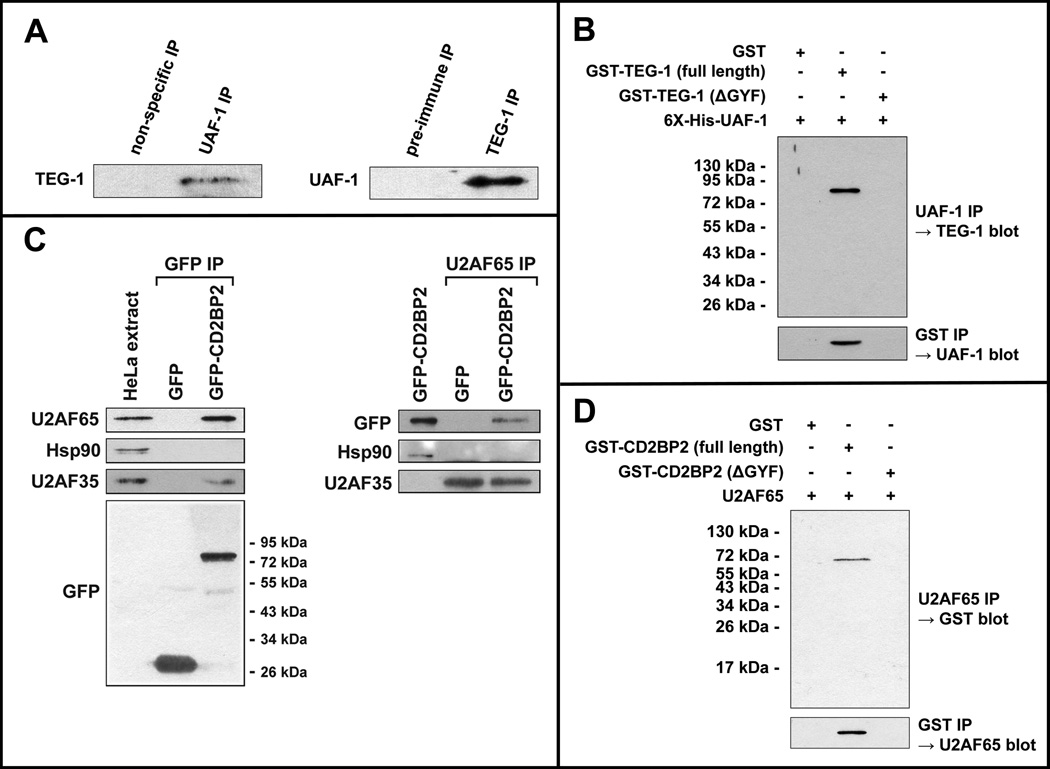
The genetic and expression analyses described above suggest that TEG-1 is functioning, at least in part, as a splicing factor. Additionally, the human and yeast TEG-1 homologs physically interact with known splicing factors (see Discussion), although the overall similarity between TEG-1 and its homologs is not overly high, so it cannot be assumed that TEG-1 would have the same binding partners in C. elegans (Figure 3). To increase our understanding of how TEG-1 may function in the splicing reaction, we sought to identify proteins with which it binds. To accomplish this we expressed a TAP-tagged version of TEG-1 in animals and immunoprecipitated (IP’d) TAP::TEG-1 and associated proteins. Importantly, this tagged version of TEG-1 rescues teg-1(oz230) mutant phenotypes, and is expressed at a similar level as endogenous TEG-1, suggesting that TAP::TEG-1 is functioning similarly to the wild-type protein (Figure 5). After TEG-1 and its associated proteins were IP’d, we subjected the samples to mass-spectometry analysis to determine the identities of the interacting proteins, and compared these proteins to control IPs. UAF-1, a conserved splicing factor that is homologous to mammalian U2AF65, was identified in two independent TAP::TEG-1 IPs, but was not detected in either of the two control IPs. A full description of proteins found to bind TEG-1 will be described elsewhere. In order to verify the TEG-1—UAF-1 interaction, we performed reciprocal IPs from whole worm lysates. UAF-1 was co-IP’d with TEG-1 (using TEG-1 specific antibodies), and TEG-1 was co-IP’d with UAF-1 (using UAF-1 specific antibodies) (Figure 6A).
Figure 6.
TEG-1 directly interacts with UAF-1 in C. elegans extracts, and this interaction is conserved in humans. (A) Reciprocal co-immunoprecipitations (IPs) were performed using UAF-1 specific antibodies (left) and TEG-1 specific antibodies (right), followed by SDS electrophoresis and western blotting. In the UAF-1 IP, TEG-1 specific antibodies detect a band of the correct size, whereas an IP using non-specific pre-immunized rabbit serum, did not result in detection of TEG-1. In the TEG-1 IP, a band of the correct size was detected with UAF-1 specific antibodies, but not when pre-immune serum was used to perform the IP. (B) E. coli expressed GST-TEG-1 and 6X-His-UAF-1 co-IP. 6X-His-UAF-1 was expressed in E. coli cells with GST, GST-TEG-1, or GST-TEG-1 with the GYF domain removed (GST-TEG-1(ΔGYF); Figure 3B). When UAF-1 specific antibodies were used to IP, only GST-TEG-1(full length) was co-IP’d (top). Likewise, when GST specific antibodies where used to IP, only the strain contain GST-TEG-1(full length) was able to co-IP UAF-1, as detected by UAF-1 specific antibodies. (C) Co-IP of U2AF65 and CD2BP2 from HeLa extracts. GFP or GFP-CD2BP2 were transiently expressed in HeLa cells and proteins were IP’d using anti-GFP specific antibodies (left) or anti-U2AF65 specific antibodies (right). U2AF65 and U2AF35 were detected in the anti-GFP IP on a western blot, while the negative control, Hsp90 heat shock protein, was not co-IP’d. Likewise, GFP-CD2BP2 was co-IP’d with U2AF65, while GFP and Hsp90 were not (right). U2AF35 was co-IP’d with U2AF65, whether using lysate from GFP expressing cells, or GFP-CD2BP2 expressing cells. (D) E. coli expressed GST-CD2BP2 and U2AF65 co-IP. U2AF65 was expressed in E. coli cells with GST, GST-CD2BP2(full length), or GST-CD2BP2(ΔGYF). Only GST-CD2BP2(full length) was co-IP’d with U2AF65. Likewise, when IP was performed using anti-GST antibodies, U2AF65 was only detected when lysate from cells expressing GST-CD2BP2(full length) was used; U2AF65 could not be detected when lysate from cells expressing GST or GST-CD2BP2(ΔGYF) was used for the IP.
These reciprocal co-IP experiments provide strong evidence that at least a proportion of TEG-1 and UAF-1 exist in a complex within the cell; however, this does not demonstrate that these two proteins directly interact. To test if the TEG-1—UAF-1 interaction is direct, we expressed both proteins in bacterial cells, where mRNA splicing does not occur and splicing factors are not present. Therefore, if TEG-1 and UAF-1 are part of the same multi-protein complex, the other proteins of the complex likely do not exist within the bacterial cell, and only direct interactions would be detected. We found that bacterially expressed TEG-1 and UAF-1 co-IP (Figure 6B), suggesting that the two proteins directly bind. Furthermore, when we expressed a truncated version of TEG-1, which lacks a potential (although quite degenerate) GYF protein binding motif (called TEG-1ΔGYF; Figure 3), an interaction between TEG-1ΔGYF and UAF-1 was not detected (Figure 6B). The lack of interaction between TEG-1ΔGYF and UAF-1 provides an important negative control, suggesting that the bacterial TEG-1—UAF-1 co-IPs are specific. Furthermore, the lack of detectable binding between TEG-1ΔGYF and UAF-1 suggests that the degenerate GYF domain is necessary for the TEG-1—UAF-1 interaction. While reducing uaf-1 activity through RNAi results in an embryonic lethal phenotype (Zorio and Blumenthal, 1999b), partially reducing uaf-1 activity through RNAi in a glp-1(gf) background results in a significant increase in proliferation in the germ line (Table 3), suggesting that, like teg-1, uaf-1 is also involved in regulating the proliferation vs. differentiation decision in the C. elegans germ line. Additionally, since a complete reduction of teg-1 activity in teg-1 null mutants still allows for animals to reach adulthood, while severely reducing uaf-1 activity results in embryonic lethality, suggests that the TEG-1 function may be modulatory.
Table 3.
Interaction of uaf-1 with glp-1(ar202gf).
| Germline Phenotype at 18°C (%) | ||||
|---|---|---|---|---|
| Genotype | Wild Type | Mog2 | Pro3 | n1 |
| rrf-14; glp-1(ar202gf); pL4440(RNAi)5 pL4440(RNAi)5 | 96 | 0 | 4 | 54 |
| rrf-1; uaf-1(RNAi)6 | 49 | 51 | 0 | 49 |
| rrf-1; glp-1(ar202gf); uaf-1(RNAi) | 32 | 15 | 53 | 34 |
Number of gonad arms
Masculinization of the germ line
Proximal proliferation. Gonads contain and over-proliferative tumor in the proximal end of the gonad.
rrf-1(pk1417)
pL4440 is an empty vector, used for control RNAi.
RNAi was performed by placing gravid hermaphrodites on RNAi plates and scoring the progeny, ensuring that progeny were not exposed to RNAi until after hatching. If uaf-1 RNAi was performed by placing L4 animals on the RNAi plate, this resulted in most of the progeny arresting as embryos, and a very small brood size.
The TEG-1—UAF-1 interaction is conserved in humans
An interaction between homologs of TEG-1 and UAF-1 has not been previously reported in any system. Given the overall low level of similarity between TEG-1 and its homologs (Figure 3), it is possible that the TEG-1—UAF-1 interaction is unique to nematodes, and not present in other systems. To test if the TEG-1—UAF-1 interaction is present in humans, we tested if the same interaction could be detected in Hela cells. A GFP-tagged version of CD2BP2, the human homolog of TEG-1, was transformed into Hela cells, followed by IP with anti-GFP antibodies (Figure 6C). As a control, a construct encoding only GFP was transformed into cells and IP’d. Using antibodies specific to U2AF65, the human homolog of UAF-1, we detected U2AF65 co-IP with GFP::CD2BP2, but not with the GFP negative control (Figure 6C). We also detected the interaction by performing the reciprocal IP; immunoprecipitate obtained using U2AF65 antibodies contained GFP::CD2BP2, as detected using anti-GFP antibodies. Additionally, CD2BP2 and U2AF65 expressed in bacterial cells were co-IP’d, suggesting that the interaction between the two proteins is direct (Figure 6D). Together, these results demonstrate that the TEG-1—UAF-1 interaction, which we first identified in C. elegans, is also present in humans.
Discussion
We isolated teg-1 alleles in two different mutant screens, each designed to identify genes that function in the proliferation vs. differentiation decision in the C. elegans germ line. We found that, in addition to its role in the proliferation vs. differentiation decision, teg-1 also functions in regulating germline sex determination, germline proliferation, and oogenesis. Cloning of teg-1 revealed that it encodes a homolog of human CD2BP2, and that it is enriched in the nucleoplasm of cells. We also demonstrated that TEG-1 physically interacts with UAF-1, the C. elegans homolog of human U2AF65. Finally, we demonstrated that this is a conserved interaction, with the human homologs of TEG-1 and UAF-1 interacting in human cells, revealing a potential new role for TEG-1 CD2BP2 in the splicing reaction.
Splicing and the balance between proliferation and differentiation
A number of genes encoding splicing factors have been identified that, when mutated, result in similar germline phenotypes in C. elegans, including masculinization of the germ line, and over-proliferation in sensitized genetic backgrounds (Puoti and Kimble, 2000; Belfiore et al., 2004; Konishi et al., 2008; Mantina et al., 2009; Kerins et al., 2010; Zanetti et al., 2011). Indeed, a comprehensive examination of splicing factors and the ability of a reduction in their activity to cause over-proliferation in different genetic backgrounds has recently been performed (Kerins et al., 2010). Of 114 splicing factors tested by RNAi, 11 disrupted germline sex determination resulting in a Mog phenotype, while 31 enhanced the over-proliferation phenotype of a glp-1(gf) mutant. Importantly, the splicing factors that resulted in these phenotypes are found throughout the various steps of the splicing reaction. Therefore, it is not just one step of the splicing reaction that can be disrupted to cause these phenotypes, nor is it likely that a subset of splicing factors have been co-opted into a function separate from the other splicing factors. Rather, it appears as though a general disruption in splicing, or the splicing machinery, results in the Mog and over-proliferation phenotypes. Attempts at identifying potential target mRNAs whose mis-splicing results in a Mog or over-proliferation phenotype have thus far been unsuccessful (Puoti and Kimble, 1999; Belfiore et al., 2004), although defects in splicing due to a reduction in one of these factors has recently been described (Zanetti et al., 2011). As we have previously suggested, a failure in detecting causative splicing defects could be due to the reduction in properly spliced product (or accumulation of mis-spliced product) being too small to detect using conventional techniques, or that the proper targets may not have yet been tested (Mantina et al., 2009). An alternate explanation could be that the reduction in the splicing machinery that causes the over-proliferation phenotype may not be due to a defect in splicing, but rather that a reduction in splicing factor activity could interfere with some other aspect of RNA metabolism, which results in over-proliferation.
Function of TEG-1/CD2BP2 in the splicing reaction
The splicing of introns from pre-mRNAs is a multistep process that involves many protein and RNA complexes. The U2 Auxiliary Factor (U2AF) is involved in the formation of the splicing commitment complex, including being necessary for proper U2 snRNP binding to the pre-mRNA (Ruskin et al., 1988; Zamore and Green, 1989). U2AF binds to the 3’ splice site of the intron, which involves U2AF65, the large subunit of U2AF, binding to a polypyrimidine tract, and the small U2AF subunit, U2AF35, binding to the AG/ splice site (Merendino et al., 1999; Wu et al., 1999; Zorio and Blumenthal, 1999a; Wahl et al., 2009; Valadkhan and Jaladat, 2010). In vertebrates, both the length of the polypyrimidine tract, and the distance between the tract and the splice site, can vary between introns (Wahl et al., 2009; Valadkhan and Jaladat, 2010). However, in C. elegans most introns have a highly conserved consensus, UUUUCAG/R, with no spacing between the tract and the splice site (Zhang and Blumenthal, 1996; Blumenthal and Steward, 1997; Kent and Zahler, 2000). Both UAF-1, the C. elegans homolog of U2AF65 (Zorio et al., 1997), and UAF-2, the C. elegans homolog of U2AF35 (Zorio and Blumenthal, 1999b), bind to this consensus sequence (Zorio and Blumenthal, 1999a), and are likely responsible for the high degree of sequence conservation in the consensus C. elegans 3’ splice site (Hollins et al., 2005).
Homologs of TEG-1 have been implicated in splicing. Human CD2BP2 co-purifies with the U5 snRNP, binding the 15 kDa and 102 kDa proteins; therefore, CD2BP2 has also been called U5-52K (Bach et al., 1989; Behrens and Luhrmann, 1991; Laggerbauer et al., 2005). During the stepwise assembly of the spliceosome, the U5 snRNP associates with the U4/U6 di-snRNP to form the U4/U6.U5 tri-snRNP (Behrens and Luhrmann, 1991). Importantly, even though CD2BP2(U5-52K) binds to the U5 snRNP, it is not found in the U4/U6.U5 tri-snRNP, suggesting that CD2BP2(U5-52K) may be involved in the assembly of the tri-snRNP, but dissociates from U5 during tri-snRNP formation. (Laggerbauer et al., 2005). The ortholog of TEG-1 in Saccharomyces cerevisiae, Lin1p, also binds to the U5 snRNP, suggesting that this role in tri-snRNP assembly is conserved (Bialkowska and Kurlandzka, 2002).
We propose that the interaction we have detected between TEG-1 CD2BP2(U5-52K) and UAF-1 U2AF65 reveals a function for TEG-1 CD2BP2(U5-52K) that is distinct from its role in tri-snRNP formation. Indeed, proteomic analyses of Complex A and Complex B support a function for TEG-1 CD2BP2 that is independent of tri-snRNP formation. The U4/U6.U5 tri-snRNP binds to the spliceosome as a preassembled complex (Bindereif and Green, 1987; Cheng and Abelson, 1987; Konarska and Sharp, 1987). However, CD2BP2(U5-52K) was detected in experiments in which Complexes A and B were isolated and their protein constituents determined through proteomic analysis (Hartmuth et al., 2002; Deckert et al., 2006). If CD2BP2(U5-52K) is only present in the U5 snRNP prior to U4/U6.U5 tri-snRNP formation, and if the tri-snRNP is pre-assembled prior to becoming part of the spliceosome, then CD2BP2(U5-52K) should never be part of the spliceosome as it is being assembled on the pre-mRNA, including as part of Complex A or Complex B. Furthermore, the U4/U6.U5 tri-snRNP has not yet become part of the splicesome during Complex A formation. Therefore, even if CD2BP2(U5-52K) does have some affinity for U5 after tri-snRNP formation, it should not be isolated with Complex A. Our identification of TEG-1 CD2BP2(U5-52K) as binding to UAF-1 U2AF65 is consistent with its presence in Complexes A and B, and suggests an additional function for TEG-1 CD2BP2(U5-52K).
We do not know when or why TEG-1 CD2BP2(U5-52K) binds to UAF-1 U2AF65 during spliceosome assembly. However, U2AF65 has been shown to bind other proteins during spliceosome assembly. For example, in both yeast and humans U2AF65 binds to the branch point binding protein SF1, presumably to aid in commitment complex formation (Abovich and Rosbash, 1997). Additionally, SF3b155, which is part of the U2snRNP, binds to U2AF65 as the U2snRNP replaces SF1 at the branch point (Gozani et al., 1998). Two-hybrid analysis also identified multiple other U2AF65 splicing factor binding partners (Prigge et al., 2009); however, the functions of these interactions are unknown. Therefore, U2AF65 binds different proteins as formation of the spliceosome progresses. TEG-1 CD2BP2(U5-52K) binding to UAF-1 U2AF65 may be another such interaction functioning in spliceosome formation. Although we do not yet know what the consequences of this interaction may be, it is intriguing that both UAF-1 U2AF65 and the U5 snRNP, with which TEG-1 CD2BP2(U5-52K) was previously shown to bind (Laggerbauer et al., 2005), associate with the 3’splice site. As mentioned, U2AF65 binds to the polypyrimidine tract near the 3’ splice site, with UAF-1 binding to the analogous UUUUCAG/R sequence adjacent to the 3’splice site in C. elegans. In mammalian cells, three proteins of the U5 snRNP replace U2AF in this region during spliceosome assembly (Chiara et al., 1997). Even though these three U5 proteins (110, 116 and 220 kDa) are not the same U5 snRNP proteins with which CD2BP2(U5-52K) interacts (15 and 102 kDa), it is possible that functions that CD2BP2(U5-52K) provides while binding to the U5 snRNP and to UAF-1 U2AF65 are related. For example, it is possible that TEG-1 CD2BP2(U5-52K) is involved in assisting both UAF-1 U2AF65 and the U5 snRNP to bind to the 3’ end of the intron. It is also possible that TEG-1 CD2BP2(U5-52K) is involved in facilitating the replacement of UAF-1 U2AF65 by the U5 snRNP. Differentiating between these and other possibilities will require studying the timing of the TEG-1/CD2BP2 interactions with UAF-1/U2AF65 and the U5 snRNP during spliceosome assembly.
Experimental Procedures
C. elegans Growth Conditions and Strains
Maintenance and manipulation of C. elegans was performed as previously described (Brenner, 1974). Bristol N2 was the wild-type strain used and the following mutants were used in this study– LGI: fog-1(q241), gld-2(q497), rrf-1(pk1417), gld-1(q485), fog-3(q443) LGII: tra-2(e2020gf), gld-3(q730), nos-3(oz231) LGIII: unc-32(e189) glp-1(ar202gf), glp-1(oz112oz120), spe-16(hc54), teg-1(oz189), teg-1(oz230), dpy-18(e364) LGIV: fem-3(e1996) LGV fog-2(q71)
Mapping teg-1
Using standard three-factor mapping, we mapped both the over-proliferation and Mog phenotypes of teg-1(oz189) to a ~0.5 cM region between, or very close to, spe-16 and dpy-18 on linkage group III. We further narrowed the critical region through a combination of single nucleotide polymorphism (SNP) mapping and deficiency mapping. The right boundary of the critical region was determined through SNP mapping using teg-1(oz189) dpy-18(e364)/HA-8. The cold-sensitive Mog phenotype was mapped by picking Dpy non Teg (Mog) animals. A recombinant was obtained that retained N2 DNA from dpy-18 to a SNP in T28D6.5, making this SNP the right boundary of the critical region (SNP between 11337877 and 11339172-wormbase freeze WS220-information provided by Kate Hill). The left boundary of the critical region was narrowed by mapping the endpoint of the deficiency tDf2, which complements teg-1(oz189). We found that tDf2 deletes part of Y47D3A.25, but not the gene immediately to the right, Y47D3A.27. Therefore, the left boundary of the critical region is in Y47D3A.25. These left and right boundaries narrowed the critical region to a portion of chromosome III containing only three genes; Y47D3A.27, Y47D3A.28 and T28D6.6. Sequencing revealed that Y47D3A.27 contained nonsense mutations corresponding to both the teg-1(oz189) and teg-1(oz230) alleles. Sequencing of the yk82g9 cDNA, as well as RT-PCR amplified cDNA (using primers R10- CCGAAACGCGCCGTTTCTTT and R11- TTACAAATAAAGCTCGAAATCA) was used to determine the inton/exon boundaries.
Generation of Anti-TEG-1 Antibodies
Two synthetic peptides, one corresponding to the TEG-1 amino-terminal sequence (CVKERPGNADEDEEEKKKFHT), and one to the carboxyl-terminal sequence (CAGAPMEQEEEEADDSVKWEY), were conjugated by Keyhole-limpet hemocyanin (KLH) and injected into rats and rabbits for antibody production by Covance (Covance Inc., Princeton, N.J.). Antibodies from each species of the antisera were affinity purified against the corresponding peptides coupled to the SulfoLink Coupling Gel (Pierce, Rockford, IL) according to the manufacturer’s instruction.
Gonad Dissections and Indirect Immunofluorescence Staining
Gonad dissections and antibody staining were performed as previously described (Jones et al., 1996). Briefly, dissected gonads were fixed with 3% paraformaldehyde for 10 min. followed by fixation/ permeabilization with −20°C, 100% methanol for > 30 min. The gonads were blocked in 3% BSA. Affinity purified rat anti-TEG-1 (N-terminal) antibodies were used at 1:500 dilution, anti-REC-8 antibodies were used at 1:200 dilution, and anti-HIM-3 antibodies (Zetka et al., 1999) were used at 1:500 dilution. Nuclear DNA was visualized by DAPI. Images were taken using a Zeiss Imager Z1 microscope equipped with an Axiocam MRM digital camera.
Plasmid Construction
TAP-tag teg-1 (pDH122) construction: The teg-1 promoter region, a 522bp fragment containing sequences downstream of rab-35 stop codon and upstream of teg-1 start codon, was PCR amplified (teg-1 Pro F primer: tctgaaaattgaacaatttgtg; teg-1 Pro R primer: tttttggctgaaaatgagtgaa) and sub-cloned into the pBluescript II KS (+) vector (Stratagene) at NotI and BamHI sites. The tandem affinity purification (TAP) tag encodes for HA-8xHis-TEV-Myc epitopes and was amplified by PCR (TAP F primer: atgtacccatacgatgtcccaga; TAP R primer:caaatcctcctcgctgatc) using the pSB_GW::TAG vector as template (Polanowska et al., 2004; Walhout and Boulton 2006). The amplified TAP-tag was then sub-cloned into the vector containing the promoter at the BamHI and EcoRI sites, where the EcoRI site was filled-in with Klenow enzyme. The teg-1 coding region was amplified, using teg-1 Gen F primer (ccgaaacgcgccgtttctt) and teg-1 Gen R primer (tgatgttgggttgcagaagcct), and sub-cloned into the EcoRV site of the pCRII-TOPO vector (Invitrogen). The sequence containing both the teg-1 promoter and the TAP-tag was sub-cloned into the KpnI and SpeI sites of this pCRII-TOPO vector, resulting in the completed rescuing construct (pDH122). pL4440-uaf-1 Construct (pDH182): The uaf-1 cDNA sequence (368–951) was sub-cloned into pL4440 vector at XbaI and HinDIII sites and used to transform the E. coli HT115(DE3) strain, which were able to produce dsRNA (Timmons and Fire, 1998).
Transgenic Array and Integration
The TAP-tag teg-1 construct (pDH122) was injected into N2 hermaphrodite gonads, together with pJM67 (an elt-2::GFP construct kindly provided by J. M. McGhee, University of Calgary) and the pRF4 plasmid as visible transformation markers (Mello et al., 1991; Fukushige et al., 1998). After following the selectable co-injection markers for two generations, the extrachromosomal arrays were randomly integrated into the genome by gamma-irradiation (137Cs) with a total dose of ~3000 rads. Two independent integrants, designated ugIs3 [tap::teg-1] (strain XB307) and ugIs4 [tap-teg-1] (strain XB308), were isolated and out crossed three times. The integrated array corresponding to ugIs3 was crossed into teg-1(oz230) and was able to rescue the sterile phenotype of teg-1(oz230) allele.
Preparation of C. elegans Extracts
To generate sufficient starting material for purification processes, proteomic analyses, and immunoprecipitation studies, the wild-type or XB307(ugIs3) transgenic line from 20 cleared, 35 mm plates were used to inoculate 250 ml S medium supplemented with a E. coli OP50 pellet concentrated from 2 L of LB broth (Sulston and Hodgkin, 1988). The worm culture was grown for 3–5 days at 20°C on a shaking platform (250 rpm) until a freshly cleared culture was generated. The live worms were harvested by sucrose gradient followed by three washes with 0.1 M NaCl (Polanowska et al., 2004). The pellet was flash frozen in liquid nitrogen and stored at −80°C. The flash frozen worm pellet was resuspended in five volumes of CSK lysis buffer (100 mM PIPES, pH 6.0, 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol (DTT), 0.3 M sucrose, 0.5% Triton X-100, 1 mM PMSF, and EDTA-free complete protease inhibitor tablet (Roche, Montreal, QC)) (Polanowska et al., 2004). The worm suspension was disrupted by French Press at 20,000 psi and the soluble lysate was obtained by centrifugation at 10,000× g for 20 min. The protein concentration was determines by Bio-Rad Protein Assay (Bio-Rad, Hercules, CA) according to the manufacturer’s instruction.
Affinity purification of TAP-TEG-1
A two-step affinity purification was used to-to prepare samples for mass spectrometry identification. To prepare pre-immune and anti-TEG-1 antibodies-coupled protein A beads, 0.5 ml of agarose protein A beads (Sigma, St. Louis, MO) were incubated with 0.5 ml of each pre-immune sera or affinity purified rabbit anti-TEG-1 (C-terminal) antibodies overnight at 4°C. The pre-immune and antibody coupled beads were cross-linked as described in (Polanowska et al., 2004). Approximately 6–8 ml of packed XB307(ugIs3) worm pellet was lysed in CSK lysis buffer as described above. The extract was affinity purified through a 2 ml Ni-NTA agarose column (Qiagen, Germantown, MD). After washing the column with five volumes of PBS containing 10 mM imidazole, the coupled proteins were eluted with 10 ml of 100 mM imidazole in PBS (137 mM NaCl, 10 mM Na2PO4-12H2O, 2.5 mM KCl, 1.8 mM KH2PO4, pH 7.2). In the second immunoaffinity step, the imidazole eluates were equally divided and batch absorbed with pre-immune- or anti-TEG-1 antibody-coupled protein A at 4°C for 2–3 hours. Beads were then transferred to a Poly-Prep column (Bio-Rad, Hercules, CA) and washed three times with 6 volumes of PBS. The bound proteins were eluted three times with 100 µl/fraction of 0.1 M glycine, pH 2.5, and then neutralized with 1 M Tris, pH 8.0. The protein samples were concentrated by speed-vac, resolved on 12.5% Laemmli SDS-PAGE gels, and then stained with Bio-Safe Coomassie (Bio-Rad, Hercules, CA). Bands were excised and samples were analyzed by the SAMS Centre for Proteomics at University of Calgary by LC-MS/MS for protein identification. Peptide hits that were found in the pre-immune control were subtracted from the anti-TEG-1 antibody data. Common proteins found between two independent affinity purified experiments were analyzed for their mRNA expression patterns using the NEXTDB in situ database to determine if they were germ line expressed (Kohara, 2001).
Co-Immunoprecipitation (Co-IP)
For UAF-1 co-IP, extracts from 1.5 ml of packed N2 pellet was obtained as described above. For each immunoprecipitation, 10 mg of the extract was incubated overnight at 4°C with of ~ 20 µg of pre-immune sera, anti-UAF-1 antibodies, or affinity purified rat anti-TEG-1 (C-terminal) antibodies coupled to protein G beads (Sigma, St. Louis, MO). The beads were washed three times with CSK lysis buffer and resuspended in 2× SDS-PAGE loading buffer. Bound proteins were analyzed by immunoblots.
Bacterial Pull-Down Experiments
TEG-1 (2–353) and UAF-1 (1–496) were sub-cloned into pGEX-4T1 (GE Healthcare, Waukesha, WI) and pET-28a (Novagen, Darmstadt, Germany), respectively. A truncated teg-1 lacking the GYF domain (amino acid residues 296–353) was also sub-cloned into pGEX-4T1. Both GST- and His6x-tag constructs were co-expressed in E. coli BL21(DE3) strain (Tolia and Joshua-Tor, 2006). For pull-down assays, His6x-UAF-1 fusion proteins that bind to GST-TEG-1 recombinant proteins were co-immunoprecipitated by immobilizing GST, GST-TEG-1, and GST-TEG-1ΔGYF on glutathione agarose beads (GE Healthcare, Waukesha, WI). On the other hand, GST recombinant proteins that interact with the His6x-UAF-1 fusion proteins were purified on Ni-NTA beads (QIAGEN, Germantown, MD). The beads were washed five times with PBS containing 0.3 M NaCl. Proteins were eluted by adding 2X SDS-PAGE loading buffer into the beads and analyzed by immunoblots.
Immunoblotting
Proteins were resolved on a 10% SDS-PAGE gel and transferred to Trans-Blot nitrocellulose membrane using Mini-Trans Blot Cell (Bio-Rad). The membrane was blocked with 5% Carnation milk and incubated overnight at 4 °C with either: (1) affinity purified TEG-1 (N-terminal) antibodies at 1:1000; (2) anti-UAF-1 antibodies (Zorio and Blumenthal, 1999) at 1:20,000; (3) anti-U2AF65 antibodies (Sigma; St. Louis, MO) at 1:2000; (4) anti-U2AF35 antibodies (Novus Biologicals; Littleton, CO) at 1:2000; (5) anti-GFP (Rockland; Gilbertsville, PA) at 1:2000; (6) anti-Hsp90α (Stressgen; San Diego, CA) at 1:1000; or (7) anti-paramyosin (MH16, hybridoma cell line from Developmental Studies Hybridoma Bank, University of Iowa) tissue culture supernatant at 1:1, in TBS (137 mM NaCl, 25 mM Tris-HCl, pH 7.2, 2.5 mM KCl)-5% milk solution. The membrane was incubated 2 hours at room temperature with HRP-conjugated secondary antibodies (Jackson Laboratories) diluted at 1:5,000 and then visualized by SuperSignal WestPico substrate (Thermo Scientific, Waltham, MA) or Amersham ECL Advance kit (GE Healthcare, Waukesha, WI). Images were collected on Kodak BioMax MR films.
HeLa Tissue Culture and Transfection
HeLa-CCL2 cell line (a gift from M. J. Lohka, University of Calgary) was grown in Dulbecco’s modified Eagle’s medium supplemented with 3.7 g/ml NaHCO3, pH 7.2, 10% fetal bovin serum, 4 mM L-glutamine, 75 µg/ml Penicillin and Streptomycin at 37°C with 5% CO2. The plasmids expressing GFP-CD2BP2 (Kofler et al., 2004) were prepared by Qiagen EndoFree maxi plasmid kit (QIAGEN, Germantown, MD) and transfected into HeLa-CCL2 using Lipofectamine 2000 (Invitrogen). At 24 hours after transfection, cells were washed three times with ice-cold PBS and lysed in HEPES lysis buffer (40 mM HEPES, pH 7.5, 120 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 mM pyrophosphate, 50 mM NaF, 1.5 mM Na3VO4, and protease inhibitor cocktail (Roche, Montreal, QC)) on ice for 30 min. with intermittent vortexing. Whole cell lysate was centrifuged at 10,000× for 10 min. at 4° C and the protein concentration of the soluble fraction was determined. For each immunoprecipitation, 2 mg of the extract was pre-cleared with the protein A beads and then incubated overnight at 4° C with 20 µg of either anti-U2AF65 monoclonal antibodies (Sigma, St. Louis, MO) or anti-GFP antibodies (Rockland Immunochemicals, Gilbertsville, PA) that were coupled to protein A beads. The beads were subsequently washed five times with the lysis buffer and dissolved in 2X SDS-PAGE loading buffer for immunoblotting analysis.
Bullet points.
-
-
TEG-1 inhibits stem cell proliferation in the C. elegans germ line
-
-
TEG-1 likely functions as a splicing factor
-
-
TEG-1 CDBP2 physically interacts with UAF-1 U2AF65
Acknowledgements
We thank Kate Hill for providing valuable information on single nucleotide polymorphisms in the teg-1 region. We thank Pavel Pasierbek and Joseph Loidl for anti-REC-8 antibodies, Monique Zetka for anti-HIM-3 antibodies and Tom Blumenthal for UAF-1 antibodies and advice. We thank Ted Hansen for generating the two TEG-1 specific peptides, Christian Freund for providing the GFP::CD2BP2 construct, Adrian Krainer for the pET9c-U2AF65 construct, and Carrie Shemanko for anti-Hsp90α antibodies and use of her tissue culture facility. We thank members of the Hansen lab, and anonymous reviewers, for critical reading of the manuscript. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). This work was funded by a grant from the National Institutes of Health (NIH Grant GM63310) to TS, and grants from the Canadian Institutes of Health Research (CIHR) and the Natural Sciences and Engineering Research Council of Canada (NSERC) to DH.
References
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- Bach M, Winkelmann G, Luhrmann R. 20S small nuclear ribonucleoprotein U5 shows a surprisingly complex protein composition. Proc Natl Acad Sci U S A. 1989;86:6038–6042. doi: 10.1073/pnas.86.16.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton MK, Kimble J. fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics. 1990;125:29–39. doi: 10.1093/genetics/125.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens SE, Luhrmann R. Immunoaffinity purification of a [U4/U6.U5] tri-snRNP from human cells. Genes Dev. 1991;5:1439–1452. doi: 10.1101/gad.5.8.1439. [DOI] [PubMed] [Google Scholar]
- Belfiore M, Pugnale P, Saudan Z, Puoti A. Roles of the C. elegans cyclophilin-like protein MOG-6 in MEP-1 binding and germline fates. Development. 2004;131:2935–2945. doi: 10.1242/dev.01154. [DOI] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- Berry LW. Department of Genetics. St. Louis: Washington University School of Medicine; 1998. Regulation of the Mitotic/Meiotic cell fate decision in Caenorhabditis elegans; p. 327. [Google Scholar]
- Berry LW, Westlund B, Schedl T. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development. 1997;124:925. doi: 10.1242/dev.124.4.925. [DOI] [PubMed] [Google Scholar]
- Bialkowska A, Kurlandzka A. Proteins interacting with Lin 1p, a putative link between chromosome segregation, mRNA splicing and DNA replication in Saccharomyces cerevisiae. Yeast. 2002;19:1323–1333. doi: 10.1002/yea.919. [DOI] [PubMed] [Google Scholar]
- Bindereif A, Green MR. An ordered pathway of snRNP binding during mammalian pre-mRNA splicing complex assembly. The EMBO journal. 1987;6:2415–2424. doi: 10.1002/j.1460-2075.1987.tb02520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T, Steward K. RNA processing and gene structure. In: Riddle DL, Blumenthal T, Meyer B, Priess JR, editors. C. elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 117–145. [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SC, Abelson J. Spliceosome assembly in yeast. Genes & development. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Chiara MD, Palandjian L, Feld Kramer R, Reed R. Evidence that U5 snRNP recognizes the 3' splice site for catalytic step II in mammals. EMBO J. 1997;16:4746–4759. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S, Kodoyianni V, Bosenberg M, Friedman L, Kimble J. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H) Development. 1996;122:1373–1383. doi: 10.1242/dev.122.5.1373. [DOI] [PubMed] [Google Scholar]
- Crittenden SL, Troemel ER, Evans TC, Kimble J. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development. 1994;120:2901. doi: 10.1242/dev.120.10.2901. [DOI] [PubMed] [Google Scholar]
- Deckert J, Hartmuth K, Boehringer D, Behzadnia N, Will CL, Kastner B, Stark H, Urlaub H, Luhrmann R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Molecular and cellular biology. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Doniach T. Activity of the sex-determining gene tra-2 is modulated to allow spermatogenesis in the C. elegans hermaphrodite. Genetics. 1986;114:53–76. doi: 10.1093/genetics/114.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle TG, Wen C, Greenwald I. SEL-8, a nuclear protein required for LIN-12 and GLP-1 signaling in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2000;97:7877–7881. doi: 10.1073/pnas.97.14.7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann C, Crittenden SL, Suh N, Kimble J. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics. 2004;168:147. doi: 10.1534/genetics.104.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann CR, Kraemer B, Wickens M, Kimble J. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev Cell. 2002;3:697–710. doi: 10.1016/s1534-5807(02)00322-2. [DOI] [PubMed] [Google Scholar]
- Ellis R, Schedl T. Sex determination in the germ line (April 4, 2006) In: WormBook, editor. The C. elegans Research Community. WormBook; 2006. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RE, Kimble J. The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics. 1995;139:561–577. doi: 10.1093/genetics/139.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PM, Vought VE, Hanazawa M, Lee MH, Maine E, Schedl T. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development. 2011;138:2223–2234. doi: 10.1242/dev.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Barton MK, Kimble J, Schedl T. gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics. 1995a;139:579–606. doi: 10.1093/genetics/139.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Maine E, Schedl T. Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics. 1995b;139:607–630. doi: 10.1093/genetics/139.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund C, Dotsch V, Nishizawa K, Reinherz EL, Wagner G. The GYF domain is a novel structural fold that is involved in lymphoid signaling through proline-rich sequences. Nat Struct Biol. 1999;6:656–660. doi: 10.1038/10712. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Hawkins MG, McGhee JD. The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Developmental biology. 1998;198:286–302. [PubMed] [Google Scholar]
- Gallegos M, Ahringer J, Crittenden S, Kimble J. Repression by the 3' UTR of fem-3, a sex-determining gene, relies on a ubiquitous mog-dependent control in Caenorhabditis elegans. Embo J. 1998;17:6337–6347. doi: 10.1093/emboj/17.21.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PL, Kimble J. The mog-1 gene is required for the switch from spermatogenesis to oogenesis in Caenorhabditis elegans. Genetics. 1993;133:919–931. doi: 10.1093/genetics/133.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PL, Schedl T, Kimble J. More mog genes that influence the switch from spermatogenesis to oogenesis in the hermaphrodite germ line of Caenorhabditis elegans. Dev Genet. 1993;14:471–484. doi: 10.1002/dvg.1020140608. [DOI] [PubMed] [Google Scholar]
- Hansen D, Hubbard EJ, Schedl T. Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev Biol. 2004a;268:342–357. doi: 10.1016/j.ydbio.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Hansen D, Schedl T. The regulatory network controlling the proliferation-meiotic entry decision in the Caenorhabditis elegans germ line. Curr Top Dev Biol. 2006;76:185–215. doi: 10.1016/S0070-2153(06)76006-9. [DOI] [PubMed] [Google Scholar]
- Hansen D, Wilson-Berry L, Dang T, Schedl T. Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development. 2004b;131:93–104. doi: 10.1242/dev.00916. [DOI] [PubMed] [Google Scholar]
- Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Luhrmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc Natl Acad Sci U S A. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994;120:2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D, Klass M. Development of the reproductive system of Caenorhabditis elegans. Dev Biol. 1976;49:200–219. doi: 10.1016/0012-1606(76)90267-0. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics. 1986;114:15–52. doi: 10.1093/genetics/114.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollins C, Zorio DA, MacMorris M, Blumenthal T. U2AF binding selects for the high conservation of the C. elegans 3' splice site. RNA. 2005;11:248–253. doi: 10.1261/rna.7221605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Verheyden JM, Kimble J. Cyclin E and Cdk2 control GLD-1, the mitosis/meiosis decision, and germline stem cells in Caenorhabditis elegans. PLoS genetics. 2011;7 doi: 10.1371/journal.pgen.1001348. e1001348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 1996;180:165. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- Jones AR, Schedl T. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 1995;9:1491–1504. doi: 10.1101/gad.9.12.1491. [DOI] [PubMed] [Google Scholar]
- Kadyk LC, Kimble J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Zahler AM. Conservation, regulation, synteny, and introns in a large-scale C. briggsae-C. elegans genomic alignment. Genome Res. 2000;10:1115–1125. doi: 10.1101/gr.10.8.1115. [DOI] [PubMed] [Google Scholar]
- Kerins JA, Hanazawa M, Dorsett M, Schedl T. PRP-17 and the pre-mRNA splicing pathway are preferentially required for the proliferation versus meiotic development decision and germline sex determination in Caenorhabditis elegans. Dev Dyn. 2010;239:1555–1572. doi: 10.1002/dvdy.22274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian DJ, Hubbard EJ. Caenorhabditis elegans germline patterning requires coordinated development of the somatic gonadal sheath and the germ line. Dev Biol. 2005;279:322–335. doi: 10.1016/j.ydbio.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol. 2007;23:405–433. doi: 10.1146/annurev.cellbio.23.090506.123326. [DOI] [PubMed] [Google Scholar]
- Kofler M, Heuer K, Zech T, Freund C. Recognition sequences for the GYF domain reveal a possible spliceosomal function of CD2BP2. J Biol Chem. 2004;279:28292–28297. doi: 10.1074/jbc.M402008200. [DOI] [PubMed] [Google Scholar]
- Kofler MM, Freund C. The GYF domain. FEBS J. 2006;273:245–256. doi: 10.1111/j.1742-4658.2005.05078.x. [DOI] [PubMed] [Google Scholar]
- Kohara Y. Systematic analysis of gene expression of the C. elegans genome. Tanpakushitsu Kakusan Koso. 2001;46:2425–2431. [PubMed] [Google Scholar]
- Konarska MM, Sharp PA. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Konishi T, Uodome N, Sugimoto A. The Caenorhabditis elegans DDX-23, a homolog of yeast splicing factor PRP28, is required for the sperm-oocyte switch and differentiation of various cell types. Dev Dyn. 2008;237:2367–2377. doi: 10.1002/dvdy.21649. [DOI] [PubMed] [Google Scholar]
- Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr Biol. 1999;9:1009–1018. doi: 10.1016/s0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- Laggerbauer B, Liu S, Makarov E, Vornlocher HP, Makarova O, Ingelfinger D, Achsel T, Luhrmann R. The human U5 snRNP 52K protein (CD2BP2) interacts with U5-102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation. Rna. 2005;11:598–608. doi: 10.1261/rna.2300805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont LB, Crittenden SL, Bernstein D, Wickens M, Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- MacDonald LD, Knox A, Hansen D. Proteasomal regulation of the proliferation vs. meiotic entry decision in the C. elegans germ line. Genetics. 2008;180:905–920. doi: 10.1534/genetics.108.091553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Villeneuve AM. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 2001;15:1674–1687. doi: 10.1101/gad.902601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantina P, Macdonald L, Kulaga A, Zhao L, Hansen D. A mutation in teg-4, which encodes a protein homologous to the SAP130 pre-mRNA splicing factor, disrupts the balance between proliferation and differentiation in the C. elegans germ line. Mech Dev. 2009;126:417–429. doi: 10.1016/j.mod.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Mazroui R, Puoti A, Kramer A. Splicing factor SF1 from Drosophila and Caenorhabditis: presence of an N-terminal RS domain and requirement for viability. RNA. 1999;5:1615–1631. doi: 10.1017/s1355838299991872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3' splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes & development. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- Nadarajan S, Govindan JA, McGovern M, Hubbard EJ, Greenstein D. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development. 2009;136:2223–2234. doi: 10.1242/dev.034603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa K, Freund C, Li J, Wagner G, Reinherz EL. Identification of a proline-binding motif regulating CD2-triggered T lymphocyte activation. Proc Natl Acad Sci U S A. 1998;95:14897–14902. doi: 10.1073/pnas.95.25.14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper AS, Killian DJ, Hubbard EJ. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics. 2003;163:115–132. doi: 10.1093/genetics/163.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petcherski AG, Kimble J. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature. 2000;405:364–368. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- Polanowska J, Martin JS, Fisher R, Scopa T, Rae I, Boulton SJ. Tandem immunoaffinity purification of protein complexes from Caenorhabditis elegans. Biotechniques. 2004;36:778–780. 782. doi: 10.2144/04365BM05. [DOI] [PubMed] [Google Scholar]
- Prigge JR, Iverson SV, Siders AM, Schmidt EE. Interactome for auxiliary splicing factor U2AF(65) suggests diverse roles. Biochimica et biophysica acta. 2009;1789:487–492. doi: 10.1016/j.bbagrm.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A, Kimble J. The Caenorhabditis elegans sex determination gene mog-1 encodes a member of the DEAH-Box protein family. Mol Cell Biol. 1999;19:2189–2197. doi: 10.1128/mcb.19.3.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A, Kimble J. The hermaphrodite sperm/oocyte switch requires the Caenorhabditis elegans homologs of PRP2 and PRP22. Proc Natl Acad Sci U S A. 2000;97:3276–3281. doi: 10.1073/pnas.97.7.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racher H, Hansen D. Translational control in the C. elegans hermaphrodite germ line. Genome. 2010;53:83–102. doi: 10.1139/g09-090. [DOI] [PubMed] [Google Scholar]
- Ruskin B, Zamore PD, Green MR. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Schedl T, Kimble J. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics. 1988;119:43–61. doi: 10.1093/genetics/119.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Sulston J, Hodgkin J. Methods. In: Wood WB, editor. The Nematode Caenorhabditis elegans. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. pp. 587–606. [Google Scholar]
- Tax FE, Yeargers JJ, Thomas JH. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature. 1994;368:150–154. doi: 10.1038/368150a0. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Tolia NH, Joshua-Tor L. Strategies for protein coexpression in Escherichia coli. Nature methods. 2006;3:55–64. doi: 10.1038/nmeth0106-55. [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Jaladat Y. The spliceosomal proteome: at the heart of the largest cellular ribonucleoprotein machine. Proteomics. 2010;10:4128–4141. doi: 10.1002/pmic.201000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Walhout AJM, Boulton SJ. Biochemistry and molecular biology. In: Community TCeR, editor. WormBook. WormBook; 2006. [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3' splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- Yochem J, Greenwald I. glp-1 and lin-12, genes implicated in distinct cell-cell interactions in C. elegans, encode similar transmembrane proteins. Cell. 1989;58:553–563. doi: 10.1016/0092-8674(89)90436-4. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Green MR. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci U S A. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti S, Meola M, Bochud A, Puoti A. Role of the C. elegans U2 snRNP protein MOG-2 in sex determination, meiosis, and splice site selection. Developmental biology. 2011;354:232–241. doi: 10.1016/j.ydbio.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Blumenthal T. Functional analysis of an intron 3' splice site in Caenorhabditis elegans. RNA. 1996;2:380–388. [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T. Both subunits of U2AF recognize the 3' splice site in Caenorhabditis elegans. Nature. 1999a;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA. 1999b;5:487–494. doi: 10.1017/s1355838299982225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Lea K, Blumenthal T. Cloning of Caenorhabditis U2AF65: an alternatively spliced RNA containing a novel exon. Mol Cell Biol. 1997;17:946–953. doi: 10.1128/mcb.17.2.946. [DOI] [PMC free article] [PubMed] [Google Scholar]