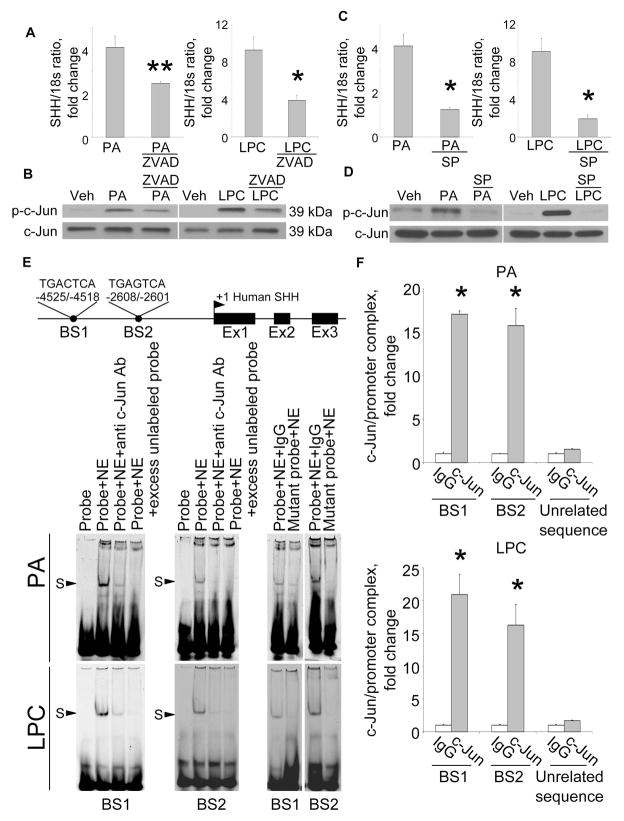
Figure 3. PA- and LPC-induce SHH expression via AP-1 mediated transcription.
(A) shC9 cells incubated with PA at 800 μM for 16 hours in the absence or presence of the pancaspase inhibitor Z-VAD-fmk (ZVAD, 10 μM) and SHH mRNA expression quantified by real-time PCR. (B) Cells were treated with vehicle (Veh), PA as above or LPC at 85 μM for 8 hrs. Immunoblot analysis was performed for phospho-c-Jun (p-c-Jun) or total c-Jun using whole cell lysates. (C) shC9 cells were treated with PA as per panel A in the presence or absence of the JNK inhibitor SP600125 (50 μM) and SHH mRNA expression quantified using real-time PCR. (D) Whole cell lysates were prepared from shC9 cells treated with PA or LPC as above and immunoblot analysis performed for p–c-Jun or total c-Jun. (E) A schematic for the SHH gene is depicted along with the two putative AP-1 binding sites (BS1 and BS2). shC9 cells were incubated with PA or LPC as described above. Nuclear protein extracts were isolated and EMSA performed using a CY 5.5-labeled AP-1 probe. Anti-phospho-c-Jun antibodies were used in the supershift experiments. Protein complexes are indicated by arrows (S, shift). No specific retarded complexes were observed when adding 200-fold molar excess of the unlabeled probe or when using the CY 5.5-labeled mutated probe. CY 5.5-labeled probe only was used as a negative control. (F) shC9 cells were incubated with PA or LPC as above. Cells were fixed and lysed, DNA fragments pulled down with p-c-Jun antibody, and the collected DNA subjected to quantitative RT-PCR analysis using primers for BS1 or BS2. DNA fragments pulled down with IgG were used as negative control for the p-c-Jun specific antisera. Primers of unrelated sequence were used as negative control for the BS1 or BS2 specific primers. All data are expressed as mean ± SEM for three experiments; *p<0.01, **p<0.05.