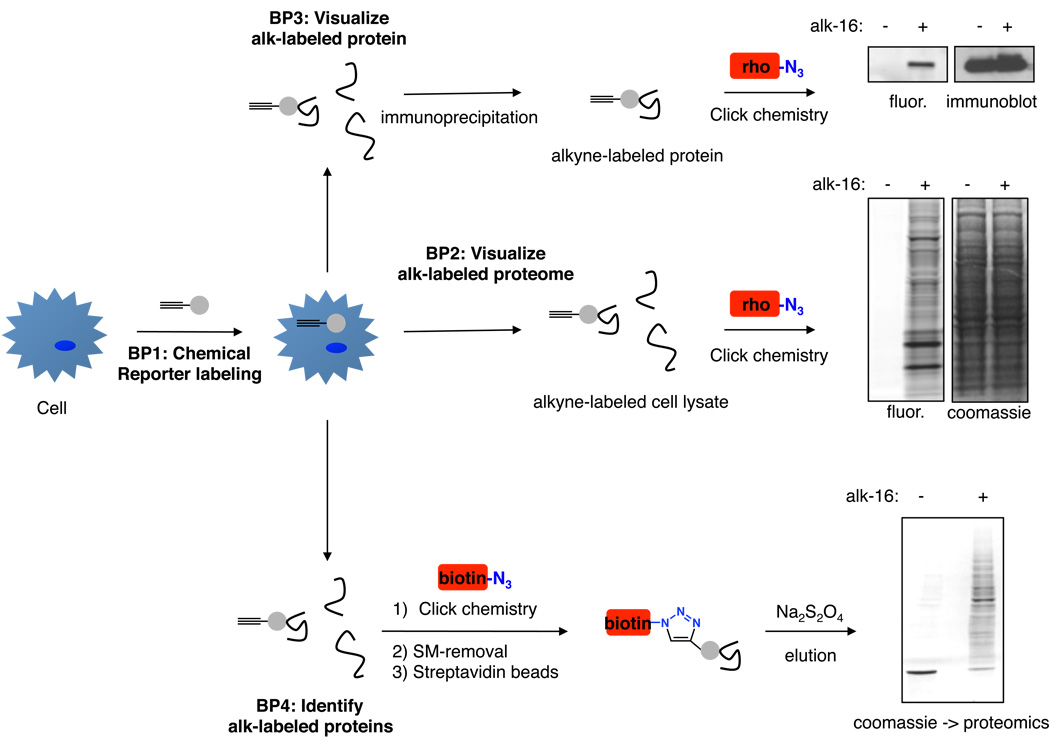
Figure 2. Workflow of protocols used for studying protein fatty acylation.
Basic protocol 1 describes labeling of proteins in cells with alkyne-chemical reporters of fatty acylation. Basic protocols 2–4 utilize the labeled cells for visualization of labeled proteomes by bioorthogonal ligation with azido-rhodamine (rho, Basic protocol 2), visualization of individual candidate proteins using immunoprecipitation and bioorthogonal ligation with azido-rhodamine (Basic protocol 3), and/or affinity enrichment of labeled proteins by bioorthogonal ligation with azido-azo-biotin (biotin) and selective elution from streptavidin beads (Basic protocol 4). Data shown for Basic protocols 2 and 4 is from DC2.4 cells labeled with 50 uM alk-16 for 1 h. Data shown for Basic protocol 3 is from HeLa cells transfected with HA-tagged IFITM3 protein and labeled with 50 uM alk-16 for 1 h followed by immunoprecipitation and immunoblotting with anti-HA antibodies.