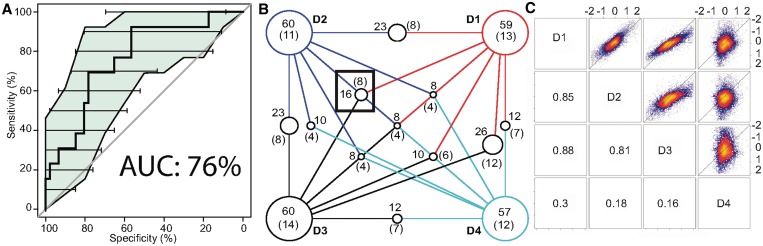
Figure 2.
Validation of OHC predictions (A) ROC curve using TFIs in BioGRID as benchmark, colored area and horizontal lines are confidence intervals of sensitivity and specificity, respectively. (B) Overlap between predicted and validated pairs between all datasets. D1: total mRNA fraction D2: labeled mRNA fraction, D3: mRNA data from Mitchell et al. and D4: Pol II ChIP-chip occupancy measurements; predictions across datasets agree well, dataset D4 having the most distinct predictions. Each intersect is shown as a circle, with radius proportional to the intersect size. The black box shows the intersect used as novel predictions. The numbers in parentheses indicate the subset of interactions that are validated by BioGRID. (C) Pairwise comparison of expression or occupancy values for all genes. Numbers in lower part indicate Spearman's correlation between datasets. D1 and D3 have the highest correlation as they are both total mRNA expression measurements. D2 has a very good but lower correlation with D1 and D3. This is due to subtle differences when measuring newly synthesized mRNA. D4 has a very weak positive correlation as Pol2 occupancy as an indicator of gene activity is very different from mRNA expression.