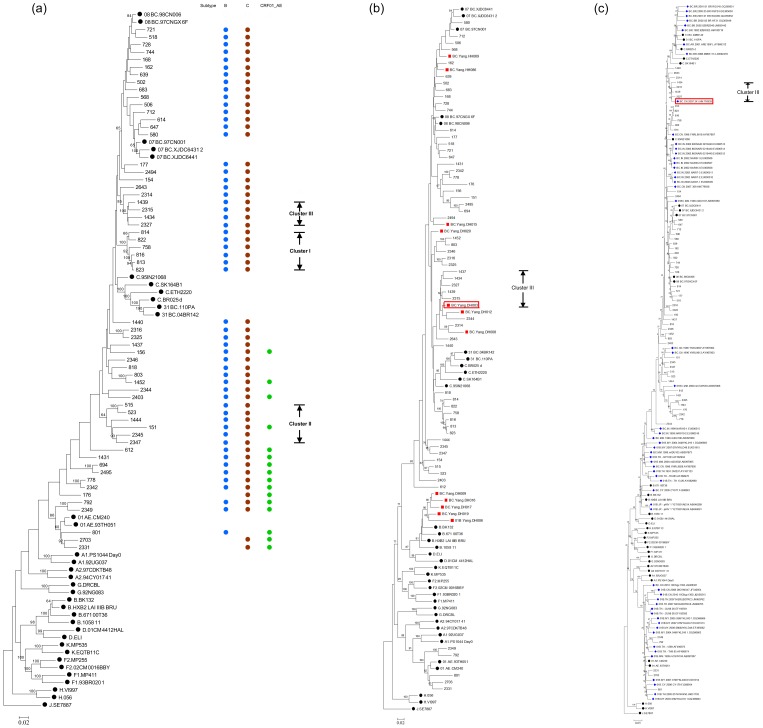
Figure 1. Phylogenetic tree analysis.
(a) Full length gag and pol genes from same isolates were assembled together. Maximum Likelihood trees created with the 59 full length gag-pol gene of HIV-1 URFs sequences from Yunnan and a selection of reference sequences of subtype A–D, F–H, J, K, CRF01_AE, CRF07_BC, CRF08_BC and CRF31_BC (black dots, http://hiv-web.lanl.gov/- see Materials and Methods for details of selection of reference sequences). The length of sequences used in the analysis was 4307 base pairs respectively by using HXB2 as the reference genomic. Each reference sequence is labeled with the HIV-1 subtype, followed by name. Branch support values greater than 60% are indicated at the corresponding nodes of the tree. The scale bar represents 2% genetic distance. The compositions of genomes were indicated in right column. Three clusters containing more than 3 strains with high branch support values were labeled accordingly. (b) Full length gag and partial pol genes (2.6 Kb) of 12 strains from Yang's paper were aligned together with 59 strains identified in this study and subtype references sequences. ML tree was constructed as described in the methods. Strains from Yang's paper were labeled with the HIV-1 subtype, followed by “Yang” and strain's name. Branch support values greater than 60% are indicated at the corresponding nodes of the tree. All strains from Yang's paper were labeled with red square (▪). Strain located in red rectangle was the one (DH003) presenting same genomic structure as strains in Cluster III. (c) Full length gag-pol genes downloaded from HIV Los Alomas database of URFs in Asia containing B, C or CRF01_AE segments were aligned with strains identified in this study. ML tree was constructed as described in the methods. Strains downloaded from database were labeled with the HIV-1 subtype, followed by country, year, name and accession number. Branch support values greater than 60% are indicated at the corresponding nodes of the tree. All strains from database were labeled with blue diamond (♦). Strain located in red rectangle was the one (341) presenting same genomic structure as strains in Cluster III.