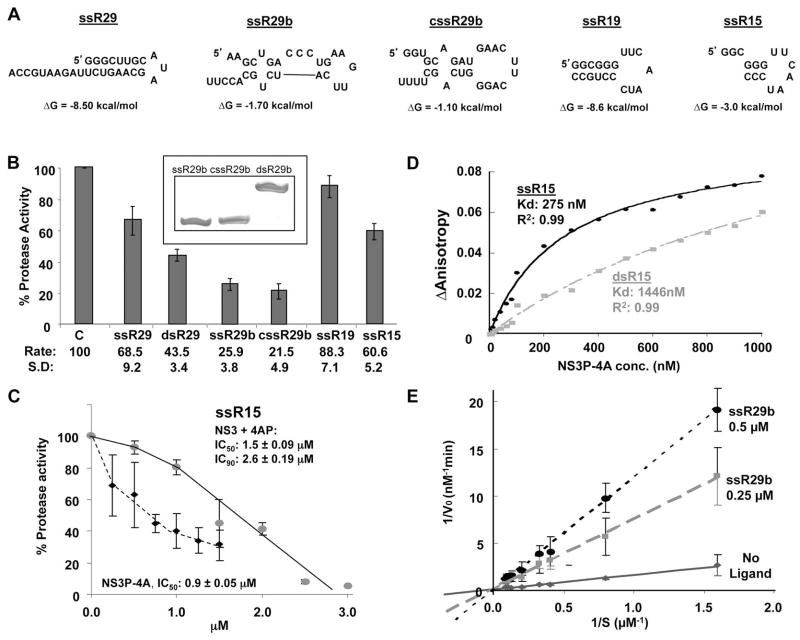
FIG. 4.
Effects of RNAs on NS3 protease activity. A) Schematics of the RNAs used in the assays. The secondary structures and predicted ΔG values of the RNAs were generated using Mfold (Zuker, 2003). B) Effects of the RNAs on NS3P-4A protease activity. Each bar represents at least three independent protease assays. The names of the RNAs used are indicated below the bars of the graph. dsR29 represents the annealed products from ssR29b and cssR29b. A demonstration that the annealing of ssR29b and cssR29b resulted in dsRNAs is shown as an inset above the bar graphs. The RNAs were electrophoresed in a nondenaturing 20% polyacrylamide gel and stained with Toluidine Blue. 100 pmoles each of the RNAs were used in the electrophoresis. C) The effects and IC50/90 of the best RNA inhibitor in panel B, ssR29b, on inhibition of NS3-4A or NS3P-4A protease activities. D) Affinities of NS3P-4A for RNAs ssR15 and dsR15. ssR15 was chemically synthesized to have a fluoresceine at the 5′ terminus and then added to a fluorescence anisotropy assay performed in an HP LS55 fluorimeter. The same assay was also performed with the labeled ssR15 annealed to the complementary RNA to generate dsR15. E) Analysis of the mechanism of inhibition of ssR29b on NS3P-4A protease activity. A standard NS3P-4A protease activity assay was performed in the presence of 4 μM substrate. The effects of two concentrations of ssR29b on the rates of cleavage by NS3P were graphed in a double reciprocal plot. The intercept for the inhibitors was approximately at zero, suggesting that the RNA antagonists for NS3P-4A were competing with the peptide substrate.