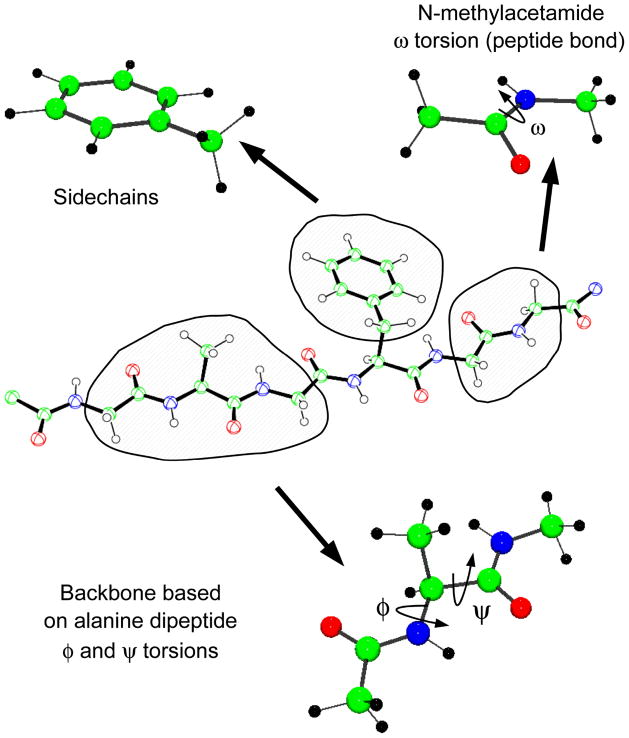
Figure 3.
Illustration of model compounds used in the parametrization of the CHARMM additive protein force field. (a) N-methyl acetamide is used to model the peptide bond; (b) sidechains, such as in PHE, are modeled by analogous compounds that include terminating methyl or ethyl groups; (c) alanine dipeptide is the model compound for optimization of the ϕ/ψ torsional parameters including CMAP corrections.