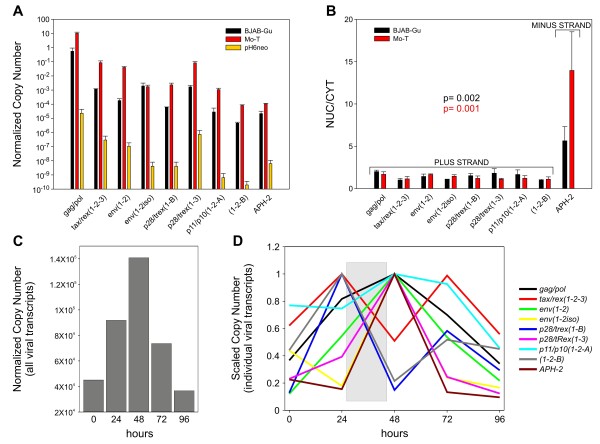
Figure 3.
Quantitative analysis, intracellular compartmentalization and expression kinetics of HTLV-2 mRNAs in infected cell lines. Normalized copy number (NCN) and Scaled Copy Numbers (SCN) were calculated as described in the text. Housekeeping genes used for normalization were GAPDH (panels A and B) and 18S rRNA (panels C and D) and in this latter case (panel C) the obtained NCN were multiplied for 107. A. NCN of viral mRNAs in the HTLV-2-infected cell lines BJAB-Gu (black bars) and Mo-T (red bars) and in cells transfected with the HTLV-2A molecular clone pH6neo (yellow bars). Cells were harvested at densities ranging from 100,000/ml (transfected cells) to 500,000/ml (infected cell lines). Shown are mean values of 3 independent experiments and standard error bars. B. Nucleo-cytoplasmic distribution (NUC/CYT) of plus- and minus-strand HTLV-2 mRNAs in infected cell lines Mo-T (red bars) and BJAB-Gu (black bars) was determined as detailed in Additional file 1: Table S1. Shown are mean values of 4 independent experiments, standard error bars and P values. The square brackets in the figure indicate a statistically significant difference between plus-and minus-strand transcripts calculated with a Mann–Whitney Rank Sum Test (p = 0.001 and p = 0.002 for the Mo-T and BJAB-Gu cell lines respectively). C,D. Temporal analysis of HTLV-2 expression. A confluent culture of BJAB-Gu cells was diluted to 25,000 cells/ml and aliquots were harvested every 24 h over a 96-h period. Shown are NCN of the sum of all (plus and minus strand) viral transcripts at each time point (C) and SCN of individual mRNAs (D). The grey box in the figure (panel D), highlights the switch in the pattern of expression from early to late transcripts.