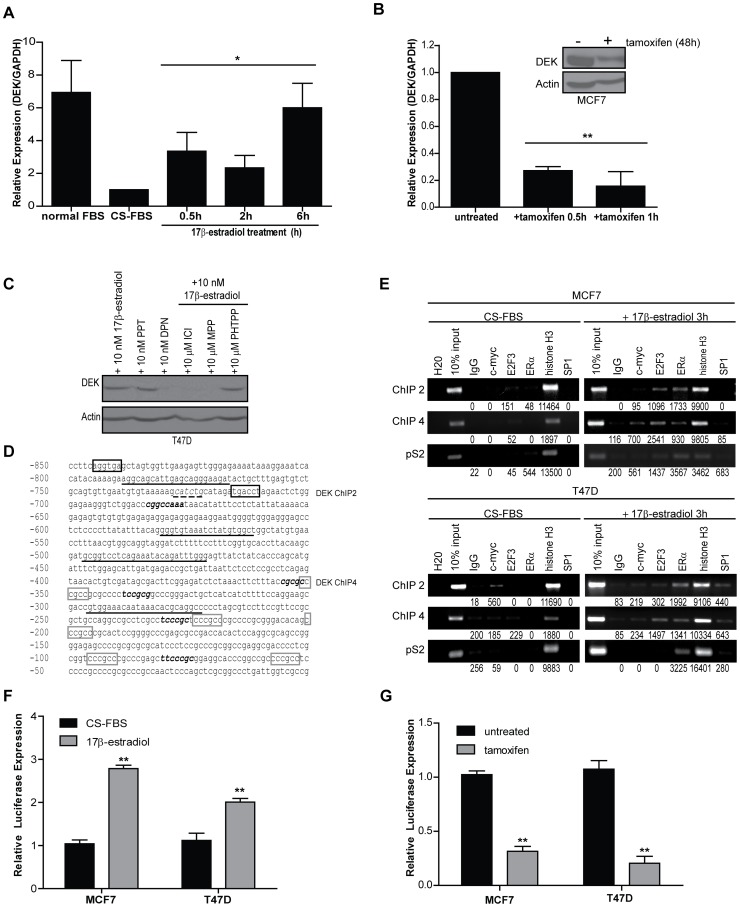
Figure 2. ERα binds to the DEK promoter in response to 17β-estradiol exposure and receptor activity correlates with DEK expression.
(A) 17β-estradiol treatment causes a rapid 4-fold increase in DEK transcription. Quantitative RT-PCR was performed to detect DEK expression in hormone starved MCF7 cells treated with 10 nM 17β-estradiol during the time course shown. GAPDH was used as a control and values are normalized to the untreated sample and represented as fold-change. Results shown depict the average of two to five independent experiments. An asterisk (*) indicates p≤0.05 as determined by one-way ANOVA. (B) Treatment with 3 µg/ml tamoxifen results in a dramatic decrease in DEK expression. Quantitative RT-PCR was performed to detect DEK expression in MCF7 cells grown in low serum treated with 3 µg/ml tamoxifien during the time course shown. GAPDH was used as a control and values are normalized to the untreated sample and represent fold change. Results shown depict the average of replicate experiments. Two asterisks (**) indicates p≤0.01 as determined by one-way ANOVA. (C) ERα, not ERβ, stimulates 17β-estradiol induced DEK expression as determined by western blot analysis. T47D cells were grown in CS-FBS for 7 days then treated with either 10 nM of agonists or 10 µM of antagonist in the presence of 10 nM 17β-estradiol. (D) DEK gene promoter −850 bp upstream of the 5′ UTR. Primers used for ChIP are depicted by underlined sequences. The ERE half sites, described as “DEK ChIP 2,” are highlighted in black boxes at positions −845 and −716 bp. The putative ERα/SP1 binding sites are highlighted with gray boxes drawn starting at position −352 and were amplified as “DEK ChIP 4.” E2F and E2F3 binding sites, previously characterized by Carro et al [27], are indicated by bold, italicized text. The putative c-myc binding site is underlined with a dashed line. (E) ERα binds to the DEK promoter in response to 17β-estradiol treatment. Chromatin was isolated from MCF7 and T47D cells cultured in CS-FBS that were either untreated or treated for 3 hours with 10 nM 17β-estradiol. Chromatin was then subjected to immunoprecipitation using antibodies for IgG (negative control), c-myc, E2F3, ERα, histone H3 (positive control), or SP1. “Input” represents 10% of the DNA used in the immunoprecipitation. Two loci were tested in the DEK promoter, “DEK ChIP 2″ and “DEK ChIP 4″ (see above). The pS2 gene promoter was used as a positive control for ERα binding following 17β-estradiol treatment. Raw densitometry values are indicated under the gel images. (F) DEK reporter assays show transcriptional up-regulation in response to 17β-estradiol treatment. MCF7 and T47D cells were transfected with a luciferase reporter construct under the control of a 1200 bp fragment of the DEK promoter and the first exon. Cells were treated with 10 nM 17β-estradiol for 24 hours. Data represent the average fold induction of luciferase expression above untreated (CS-FBS) levels from triplicate experiments. Two asterisks (**) indicate p≤0.01 as determined by Student’s t-test. (G) DEK reporter assays show transcriptional down-regulation in response to tamoxifen treatment. MCF7 and T47D cells were transfected with a luciferase reporter construct as in (F). Cells were treated with tamoxifen for 24 hours. Data represent the average fold reduction of luciferase expression below untreated levels from triplicate experiments. Two asterisks (**) indicate p≤0.01 as determined by Student’s t-test.