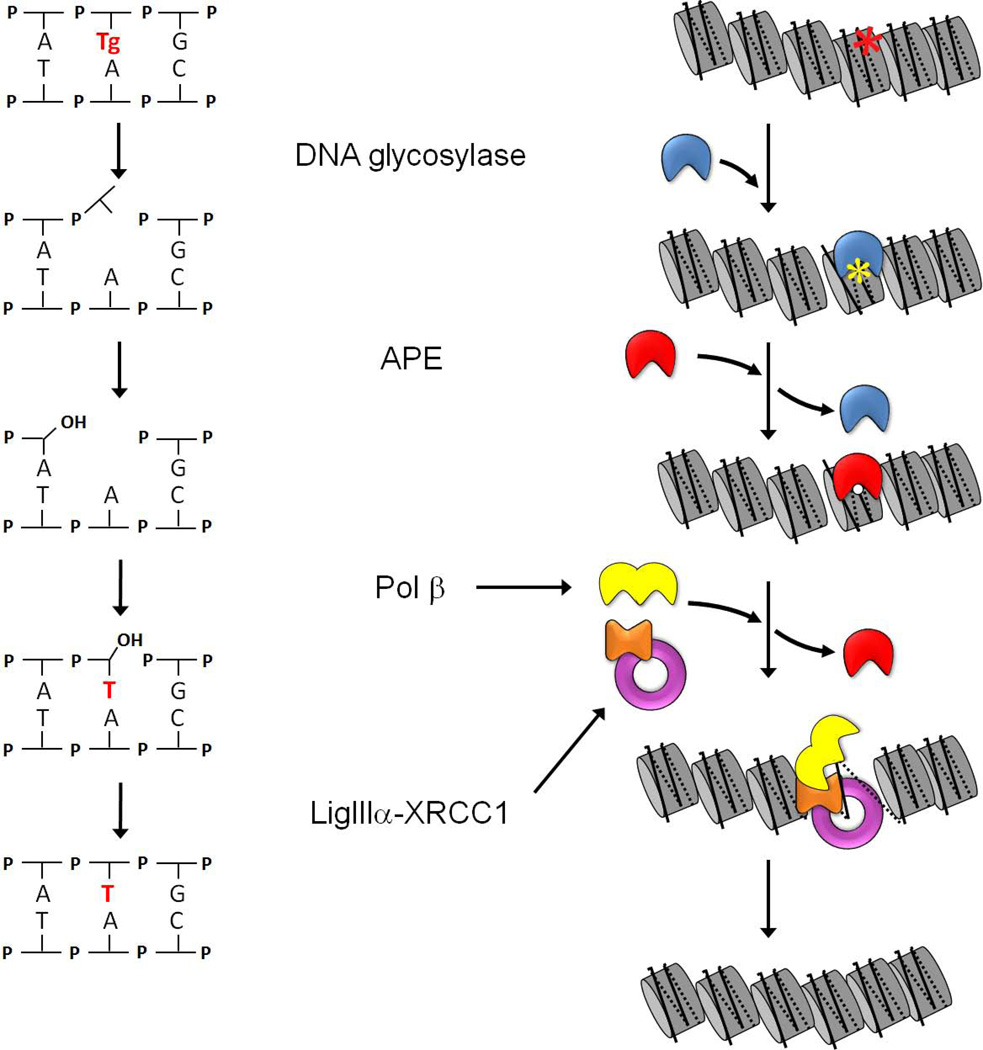
Figure 1. Short-patch base excision repair (BER; left) and a working model of BER in chromatin (right).
The left-hand schematic illustrates the enzymatic steps in short-patch BER, and the right-hand schematic depicts the corresponding steps in chromatin. Repair begins with excision of an oxidized base (in this case, thymine glycol) by a bifunctional DNA glycosylase (blue), followed by cleavage of the phosphodiester bond. APE (red) displaces the product-bound DNA glycosylase and removes the 3’ blocking moiety, leaving a gap that can be filled by DNA polymerase β (yellow) and sealed by DNA ligase IIIα (purple). The scaffolding protein XRCC1 (orange) contains separate binding sites for Pol β and DNA ligase IIIα. The complex of XRCC1 with DNA ligase IIIα disrupts DNA gap- and nick-containing nucleosomes and thus, is able to facilitate both the polymerization and ligation steps. Although not yet documented, the model depicts the re-assembly of the newly repaired DNA into a nucleosome, with the aid of histone chaperones.