Abstract
We recently presented the case of a first century AD young woman, found in the archaeological site of Cosa, showing clinical signs of malnutrition, such as short height, osteoporosis, dental enamel hypoplasia and cribra orbitalia, indirect sign of anemia, all strongly suggestive for celiac disease (CD). However, whether these findings were actually associated to CD was not shown based on genetic parameters. To investigate her human leukocyte antigen (HLA) class II polymorphism, we extracted DNA from a bone sample and a tooth and genotyped HLA using three HLA-tagging single nucleotide polymorphisms for DQ8, DQ2.2 and DQ2.5, specifically associated to CD. She displayed HLA DQ 2.5, the haplotype associated to the highest risk of CD. This is the first report showing the presence of a HLA haplotype compatible for CD in archaeological specimens.
Keywords: Celiac disease, Human leukocyte antigen haplotype, Ancient DNA, Single nucleotide polymorphisms, Malabsorption
INTRODUCTION
In 2008, we were involved in the “case of Cosa”, a skeleton of a young woman dating to the first century AD, found in the archaeological site of Cosa, southwest of Tuscany, Italy (Figure 1)[1]. Based on the physical anthropology description[2], she was a 18-20 year -old woman, dead in physical impairment, showing signs of failure to thrive and malnutrition, all signs of typical celiac disease (CD), in particular, she was slightly built and moderately short for her age (140 cm in height), with clear signs of bone fragility and osteoporosis. She showed on her orbital roof a pathological sign, the "cribra orbitalia" (yet published)[1], a bone porosity also well distinguished in the bone of the skull vault. This condition is generally linked to bone marrow hypertrophy following anemic conditions, such as iron deficient chronic anemia. This excessive porosity could be also found in the external surface of the bones, in particular in the skull vault (Figure 2A). Furthermore she showed the bone marrow reactive hypertrophy associated to bone atrophy (Figure 2B). Although teeth structure and number were normal, she presented basal dental enamel hypoplasia (Figure 2C), a marker of nutritional or infectious stress. Measuring in the femur the angle between the neck and the diaphysis, it appears larger than normal adult angle (135° vs 125°), consistent with a diagnosis of coxa valga, typical of the hip subluxation, due to congenital dysplasia. This diagnosis is supported by the flattening of the postero-superior part of the acetabular cavity. All these signs, taken together, strongly suggested an advanced state of chronic malnutrition, consistent with a typical form of CD[3-5]. From the beginning, a poor availability of food was excluded as several signs indicated that she was member of a wealthy family, as suggested by the jewels she wore and by the overall quality of her tomb. Based on ethologic data[6], we considered that her diet was variegated and probably rich in wheat, and consequently in gluten. We hypothesized that the young woman suffered from CD. Moreover, her death was caused by severe malnutrition or by a complication of it.
Figure 1.
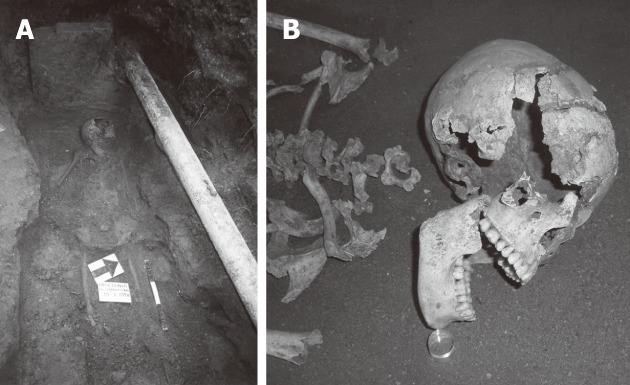
A particular of the skeleton of Cosa in the original site. A: The original site: The ancient remains of Cosa in the original site where they were found; B: The young girl: A particular of the skeleton of the young girl of Cosa.
Figure 2.
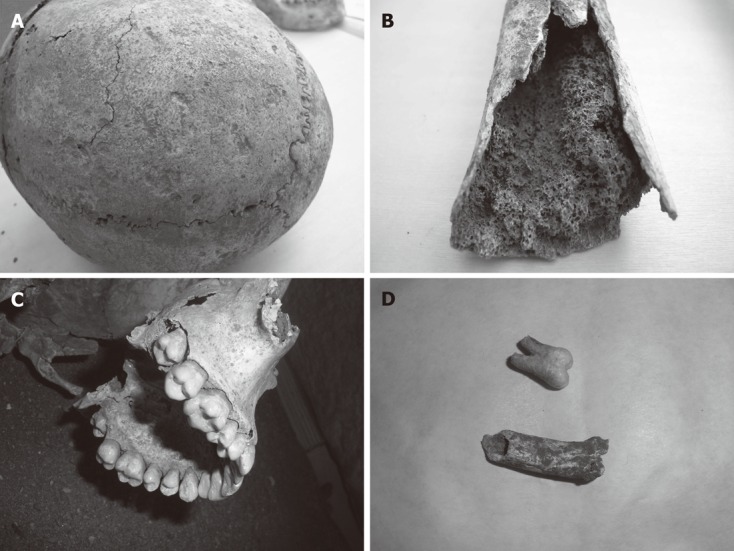
Research on the skeleton of the young girl. A: Skull vault showing excessive porosity; B: A particular of the femur showing cortical atrophy and spongiosum bone hypertrophy, indirect sign of bone marrow hypertrophy, well preserved in 2000 years; C: A detail of teeth, showing basal dental enamel hypoplasia; D: DNA extraction: A tooth and a sample of bone from which DNA was extracted.
The most ancient case of CD ever described was reported by Areteus of Cappadocia in the 1st-2nd centuries AD[7], indicating that CD could have a old origin. However the appearance of the predisposing haplotypes in humans as well as the origin of CD is still unknown.
Genetic susceptibility is a crucial step in the pathogenesis of CD, as demonstrated by studies on monozygotic twins, that show disease concordance rate of 75% compared with 11% in dyzigotic twins[8]. Furthermore siblings have an increased risk of CD, of which about 40% depends on human leukocyte antigen (HLA) genes[9]. The role of HLA class II molecules as the major genetic risk factor for CD is well known, even if the genetic effect attributable to HLA is only 54%[10]. It has been reported that over 90% of CD patients express HLA-DQ2 heterodimer while the others express HLA-DQ8[11]. In Europe only fewer than 0.5% of celiac patients express neither DQ2 nor DQ8[12], making HLA genotyping a marker with a very powerful negative predictive value, excluding the diagnosis in people who do not have CD risk HLA haplotype. Until present no information were available on CD genetics in ancient time, since DNA analysis of human remains is technically difficult.
CASE REPORT
In order to confirm the clinical hypothesis that the young woman was actually suffering from CD, we extracted DNA from parts of the skeleton and studied HLA polymorphisms known to be involved in susceptibility to CD. In particular, we analyzed DQ2 (encoded by the DQA1* 05 and DQB1*02 alleles) and DQ8 (encoded by the DQA1*03 and DQB1*0302 alleles) HLA heterodimers[13,14].
Ancient DNA analysis of human remains is particularly challenging, therefore every attempt was made to ensure the generation of authentic and meaningful data following the strictest available criteria[15-19]. To extract the genetic material, a bone sample and the left mandibular third molar (Figure 2D) were chosen since they are the best sources of DNA. Bones and teeth consist of hard material that contains small hollow spaces with single cells that are less affected by diagenetic processes and by natural contamination (microorganisms, fungus), and modern contaminations are likely to be removed prior to extraction. Bone and tooth sample were firstly brushed and irradiated for 1 h under ultraviolet (UV) light. Afterwards, the entire surface was removed by using dental drills, and the samples were cut into smaller pieces with drill. The samples were again UV-irradiated for 45 min, grounded to fine powder and stored until use at 4 °C. Only commercially certified DNA/RNA-free consumables were utilized and all tools and containers used were sterile and DNA-free. All workers wear gloves, safety masks, disposable coveralls, plus particular shoes. Every item entering is extensively washed with bleach and subsequently UV-irradiated.
As first step, we studied the preservation of bone collagen, one of the best indicator of bone preservation and therefore of DNA survival[16]. Collagen was extracted from a small bone fragment following the procedure reported in Craig et al[20]. The results obtained indicated the good state of preservation of the collagen. In fact, collagen yield expressed as weight percentage was 1.065% and the ratio C/N was in the range between 3.25 and 3.34 as expected when organic substances are saved from decay[21]. Based on these results, DNA extraction was performed in a laboratory physically isolated from all other laboratories which offers all the state-of-the-art facilities for aDNA studies[17,22,23]. They consist of a contamination resistant facility, which are maintained at positive pressure, frequently cleaned with HCl, NaClO and DNAzap™, UV and high-efficiency particulate arresting filtered, and have restricted access, designed to minimize the possibility of contamination with extant human DNA[23]. The laboratory has consecutive rooms, every room is fitted with UV-C light sources (254 nm) that can be switched on and off from outside the respective lab. The first room has an entry area for changing into suitable clean room clothing. The second room has bench space for handling sampling with fine scale for weighing of samples, a dentist drill for cutting and drilling samples as well as mortars and pestles for griding samples. Another two independent labs with hoods (with internal UV-C sources and biosafety cabinets) are used for DNA extraction and one other for polymerase chain reaction (PCR) setup.
Briefly five hundred milligrams of powder were digested in a proteinase K lysis buffer and DNA was extracted through silica-based spin columns[24]. At least two independent DNA extractions were performed on bone and teeth respectively; mock-extraction controls were carried out identically to those on the samples.
Three HLA-tagging single nucleotide polymorphism (SNPs) were genotyped in order to capture the DQ8, DQ2.2 and DQ2.5 HLA types as reported by Monsuur et al[25],
using TaqMan chemistry and the on demand assays by Applied Biosystems (Foster City, CA, United States, www.appliedbiosystems.com: Assay IDC_29817179_10dbSNP ID rs7454108; Assay IDC_29315313_10dbSNP ID rs7775228; and Assay IDC_ 58662585_10dbSNP ID rs2187668). Negative controls for amplification (PCR without template DNA) were set up simultaneously to detect contamination and at least 4 independent amplifications of each fragment were performed.
TaqMan® single nucleotide polymorphism (SNP) genotyping assays provide optimized assays for genotyping SNPs and make it easy to perform SNP genotyping discrimination studies. Samples were amplified and genotyped using the manufacturer’s instructions on an ABI Prism 7500 Fast Real Time PCR System (Applied Biosystems, Foster City, CA, United States). All SNPs were typed using the standard amplification protocol as supplied by Applied Biosystems (hold 10 min, at 95 °C, and 40 PCR cycles with denature 15 s, at 92 °C and anneal/extend 1 min, at 60 °C).
Moreover, in order to confirm the RT-PCR results, we amplified and sequenced the three predictive fragments. The list of primers designed for the experiment and the length of each PCR fragment analyzed are reported in Table 1. PCR amplification was performed in 25 μL reaction containing 2 μL DNA extract, with a final concentration of 1XPCR Gold Buffer II, 2.5 mmol MgCl2, 1 mmol dNTPs, 100 nmol primers, 0.1 mg/mL bovine serum albumin, 1 U AmpliTaq Gold (Applied Biosystems). The PCR reaction was run for 35 cycles at 94° for 30 s, 60 °C for 30 s and 72 °C for 30 s, with a first denaturation step (94 °C for 5 min), and a final extension (72 °C for 10 min).
Table 1.
Primers and length of polymerase chain reaction fragments analyzed for predicted single nucleotide polymorphism
| SNPs | Forward primer | Reverse primer | Length fragment (bp) |
| rs7454108 | ACTATTATTTCTCCAAGTTCTGACTTCCCT | GCCAAGTTGGAATAAGCCCACTATA | 155 |
| rs7775228 | AGGAAAGGAACTATCTGGGTATGGA | TGCAAAGCCCCTTTATCATTATCCT | 80 |
| rs2187668 | GTGAGGTGACACATATGAGGCAG | GGCTGAATGCCTTCAACAATCATTT | 74 |
SNPs: Single nucleotide polymorphism.
PCR products were visualized by gel electrophoresis on a 1.5% agarose gel stained with GelStar (Cambrex, Rockland, ME, United States). Positive amplification products were purified through ExoSap-IT (USB Affymetrix, Santa Clara, CA) according to manufacturer’s specifications. Afterwards, they were labeled with fluorescent dyes, purified by the ethanol precipitation technique and submitted to sequencing reaction in an ABI Prism 3100 Avant (Applied Biosystems, Foster City, CA) following the recommended sequencing kit protocols. Sequences were verified through complete overlapping of forward and reverse strands.
Genetic results were independently reproduced multiple times and all sequences were confirmed by at least two different amplified products in order to identify possible contamination.
The young girl turned out to be homozygous FAM for rs7454108, homozygous FAM for rs7775228, and homozygous VIC for rs2187668. The result is compatible with DQ2.5 homozygous genotype which is associated with higher risk of CD. This finding supports on molecular basis our hypothesis that the skeleton found in the site of Cosa suffered from CD.
Finally, to verify the endogenous nature of aDNA and track down any possible modern contaminations, molecular sex and mtDNA characterizations were performed.
Sex determination was carried out by amplification of a segment of the X-Y homologous amelogenin gene using the primer system amelogenin A/B as described by Mannucci et al[26]. This method is usually applied for typing samples of a very degraded nature, since short X and Y-specific products of 106 and 112 bp, respectively are generated from a single primer pair. The result were resolved by 12% Acrylamide electrophoresis. Molecular data confirmed the morphological and morphometric sex diagnosis of being female.
Mitochondrial DNA (mtDNA) typing[27] was performed also on the DNAs of all molecular anthropologists and archaeologists who handled the ancient sample. All the extant sequences differed from the girl "Cosa" consensus mtDNA sequence (16270T, 16362C, 73G, 150T and 263G) excluding modern DNA contamination. The ancient haplotype was certainly phylogenetically assigned to U5b2b1a haplo-group following the classification proposed by van Oven et al[28]. This haplo-group is European specific and its PAML (Phylogenetic Analysis by Maximum Likelihood) based age estimate is 9325.2 ± 3443.5 years[29-31].
DISCUSSION
This is the first report of HLA typing in ancient remains and it could be considered a very intriguing result, although it does not allow us to diagnose definitively CD. The presence of CD associated-HLA is a necessary condition, although not sufficient to develop the disease. It fact, although about 30%-35% of the actual general population express CD associated HLA genotypes, it has been estimated that, only 2%-5% of risk gene carriers develop the disease[32]. The risk increases further in homozygous for DQ2.5 (HLA-DQA1*05- DQB1*02) as shown by a recent study in United States population[12] and by another study exploring relative risks for CD in European population[33]. Another study, on sibs and parents of Italian celiac children, shows that a DQ 2.5 homozigous sib had a risk of 28% of developing CD[34]. Recent advances indicate that other genetic factors may play a role in determining which HLA compatible people could develop CD[35], particularly genes involved in T-cell regulation and inflammation, but they have not been considered for this paper. It has been reported that these genes are contributing for 3%-4% in the risk of CD, together with environmental factors, like early introduction of gluten in infants diet, early infection with enteropathic viruses or the presence of a changed bacterial flora[32].
In the case of “Cosa”, even without precise understanding of how environmental factors impacted the girl’s
life, HLA typing provide us a precious information. The presence of HLA-DQ2.5, in combination with the phenotypic observations, increases the likelihood that the young girl of Cosa suffered from CD and that CD existed already 2000 years ago, like Areteus hypothesized on clinical bases. Our results, strengthen the idea that CD was born a long time ago, walking along together with humans for a long stretch of their history, perhaps even since wheat feeding was introduced.
Footnotes
Peer reviewers: Dr. Weekitt Kittisupamongkol, Department of Medicine, Hua Chiew Hospital, 665 Bumrungmuang Road, Bangkok 10100, Thailand; Dr. Khaled Ali Jadallah, Department of Internal Medicine, King Abdullah University Hospital, Irbid 22110, Jordan; Dr. Ross McManus, Institute of Molecular Medicine, Trinity Centre for Health Science, St. James’s Hospital, Dublin 8, Ireland
S- Editor Gou SX L- Editor A E- Editor Xiong L
References
- 1.Gasbarrini G, Miele L, Corazza GR, Gasbarrini A. When was celiac disease born?: the Italian case from the archeologic site of Cosa. J Clin Gastroenterol. 2010;44:502–503. doi: 10.1097/MCG.0b013e3181d345a5. [DOI] [PubMed] [Google Scholar]
- 2.Pacciani E, Chilleri F Soprintendenza per i Beni Archeologici della Toscana. L’inumata della tomba alla cappuccina, Archeological Museum “Rovine di Cosa”, Ansedonia, Tuscany, Italy [Google Scholar]
- 3.Farrell RJ, Kelly CP. Diagnosis of celiac sprue. Am J Gastroenterol. 2001;96:3237–3246. doi: 10.1111/j.1572-0241.2001.05320.x. [DOI] [PubMed] [Google Scholar]
- 4.Tursi A, Giorgetti G, Brandimarte G, Rubino E, Lombardi D, Gasbarrini G. Prevalence and clinical presentation of subclinical/silent celiac disease in adults: an analysis on a 12-year observation. Hepatogastroenterology. 2001;48:462–464. [PubMed] [Google Scholar]
- 5.Gasbarrini G. Malabsorption syndrome. Introduction. Dig Dis. 2008;26:91. doi: 10.1159/000116764. [DOI] [PubMed] [Google Scholar]
- 6.Bellini C, Mariotti-Lippi M, Mori Secci M, Aranguren B, Perazzi P. Plant gathering and cultivation in prehistoric Tuscany (Italy) Veg Hist Archaeobot. 2008;17:Suppl 1: 103–112. [Google Scholar]
- 7.Losowsky MS. A history of coeliac disease. Dig Dis. 2008;26:112–120. doi: 10.1159/000116768. [DOI] [PubMed] [Google Scholar]
- 8.Greco L, Romino R, Coto I, Di Cosmo N, Percopo S, Maglio M, Paparo F, Gasperi V, Limongelli MG, Cotichini R, et al. The first large population based twin study of coeliac disease. Gut. 2002;50:624–628. doi: 10.1136/gut.50.5.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bevan S, Popat S, Braegger CP, Busch A, O’Donoghue D, Falth-Magnusson K, Ferguson A, Godkin A, Hogberg L, Holmes G, et al. Contribution of the MHC region to the familial risk of coeliac disease. J Med Genet. 1999;36:687–690. [PMC free article] [PubMed] [Google Scholar]
- 10.DiSabatino A, Corazza GR. Coeliac disease. Lancet. 2009;373:1480–1490. doi: 10.1016/S0140-6736(09)60254-3. [DOI] [PubMed] [Google Scholar]
- 11.Trynka G, Wijmenga C, van Heel DA. A genetic perspective on coeliac disease. Trends Mol Med. 2010;16:537–550. doi: 10.1016/j.molmed.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Pietzak MM, Schofield TC, McGinniss MJ, Nakamura RM. Stratifying risk for celiac disease in a large at-risk United States population by using HLA alleles. Clin Gastroenterol Hepatol. 2009;7:966–971. doi: 10.1016/j.cgh.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 13.Koskinen L, Romanos J, Kaukinen K, Mustalahti K, Korponay-Szabo I, Barisani D, Bardella MT, Ziberna F, Vatta S, Széles G, et al. Cost-effective HLA typing with tagging SNPs predicts celiac disease risk haplotypes in the Finnish, Hungarian, and Italian populations. Immunogenetics. 2009;61:247–256. doi: 10.1007/s00251-009-0361-3. [DOI] [PubMed] [Google Scholar]
- 14.Volta U, Villanacci V. Celiac disease: diagnostic criteria in progress. Cell Mol Immunol. 2011;8:96–102. doi: 10.1038/cmi.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper A, Poinar HN. Ancient DNA: do it right or not at all. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert MT, Bandelt HJ, Hofreiter M, Barnes I. Assessing ancient DNA studies. Trends Ecol Evol. 2005;20:541–544. doi: 10.1016/j.tree.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 17.Hofreiter M, Serre D, Poinar HN, Kuch M, Pääbo S. Ancient DNA. Nat Rev Genet. 2001;2:353–359. doi: 10.1038/35072071. [DOI] [PubMed] [Google Scholar]
- 18.Malmström H, Storå J, Dalén L, Holmlund G, Götherström A. Extensive human DNA contamination in extracts from ancient dog bones and teeth. Mol Biol Evol. 2005;22:2040–2047. doi: 10.1093/molbev/msi195. [DOI] [PubMed] [Google Scholar]
- 19.Malmström H, Svensson EM, Gilbert MT, Willerslev E, Götherström A, Holmlund G. More on contamination: the use of asymmetric molecular behavior to identify authentic ancient human DNA. Mol Biol Evol. 2007;24:998–1004. doi: 10.1093/molbev/msm015. [DOI] [PubMed] [Google Scholar]
- 20.Craig OE, Biazzo M, Colonese AC, Di Giuseppe Z, Martinez-Labarga C, Lo Vetro D, Lelli R, Martini F, Rickards O. Stable isotope analysis of Late Upper Palaeolithic human and faunal remains from Grotta del Romito (Cosenza), Italy. J Archaeol Sci. 2010;37:2504–2512. [Google Scholar]
- 21.van Klinken GJ. Bone collagen quality indicators for paleodietary and radiocarbon measurements. J Archaeol Sci. 1999;26:686–695. [Google Scholar]
- 22.Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. Genetic analyses from ancient DNA. Annu Rev Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- 23.Knapp M, Clarke AC, Horsburgh KA, Matisoo-Smith EA. Setting the stage - building and working in an ancient DNA laboratory. Ann Anat. 2012;194:3–6. doi: 10.1016/j.aanat.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Yang DY, Eng B, Waye JS, Dudar JC, Saunders SR. Technical note: improved DNA extraction from ancient bones using silica-based spin columns. Am J Phys Anthropol. 1998;105:539–543. doi: 10.1002/(SICI)1096-8644(199804)105:4<539::AID-AJPA10>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 25.Monsuur AJ, de Bakker PI, Zhernakova A, Pinto D, Verduijn W, Romanos J, Auricchio R, Lopez A, van Heel DA, Crusius JB, et al. Effective detection of human leukocyte antigen risk alleles in celiac disease using tag single nucleotide polymorphisms. PLoS One. 2008;3:e2270. doi: 10.1371/journal.pone.0002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mannucci A, Sullivan KM, Ivanov PL, Gill P. Forensic application of a rapid and quantitative DNA sex test by amplification of the X-Y homologous gene amelogenin. Int J Legal Med. 1994;106:190–193. doi: 10.1007/BF01371335. [DOI] [PubMed] [Google Scholar]
- 27.Rickards O, Martínez-Labarga C, Favaro M, Frezza D, Mallegni F. DNA analyses of the remains of the Prince Branciforte Barresi family. Int J Legal Med. 2001;114:141–146. doi: 10.1007/s004149900119. [DOI] [PubMed] [Google Scholar]
- 28.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 29.Behar DM, van Oven M, Rosset S, Metspalu M, Loogväli EL, Silva NM, Kivisild T, Torroni A, Villems R. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am J Hum Genet. 2012;90:675–684. doi: 10.1016/j.ajhg.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Available from: http: //www.familytreedna.com/ public/u5b/default. aspx? section=results.
- 31.Malyarchuk B, Derenko M, Grzybowski T, Perkova M, Rogalla U, Vanecek T, Tsybovsky I. The peopling of Europe from the mitochondrial haplogroup U5 perspective. PLoS One. 2010;5:e10285. doi: 10.1371/journal.pone.0010285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuppan D, Junker Y, Barisani D. Celiac disease: from pathogenesis to novel therapies. Gastroenterology. 2009;137:1912–1933. doi: 10.1053/j.gastro.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Margaritte-Jeannin P, Babron MC, Bourgey M, Louka AS, Clot F, Percopo S, Coto I, Hugot JP, Ascher H, Sollid LM, et al. HLA-DQ relative risks for coeliac disease in European populations: a study of the European Genetics Cluster on Coeliac Disease. Tissue Antigens. 2004;63:562–567. doi: 10.1111/j.0001-2815.2004.00237.x. [DOI] [PubMed] [Google Scholar]
- 34.Bourgey M, Calcagno G, Tinto N, Gennarelli D, Margaritte-Jeannin P, Greco L, Limongelli MG, Esposito O, Marano C, Troncone R, et al. HLA related genetic risk for coeliac disease. Gut. 2007;56:1054–1059. doi: 10.1136/gut.2006.108530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Romanos J, van Diemen CC, Nolte IM, Trynka G, Zhernakova A, Fu J, Bardella MT, Barisani D, McManus R, van Heel DA, et al. Analysis of HLA and non-HLA alleles can identify individuals at high risk for celiac disease. Gastroenterology. 2009;137:834–840, 840.e1-3. doi: 10.1053/j.gastro.2009.05.040. [DOI] [PubMed] [Google Scholar]


