Abstract
Background. Colorectal cancer (CRC) is one of the most common cancers in the world, identification of biomarkers for early detection of CRC represents a relevant target. The present study aims to determine serum peptidome patterns for CRC diagnosis. Methods. The present work focused on serum proteomic analysis of 32 health volunteers and 38 CRC by ClinProt Kit combined with mass spectrometry. This approach allowed the construction of a peptide patterns able to differentiate the studied populations. An independent group of serum (including 33 health volunteers, 34 CRC, 16 colorectal adenoma, 36 esophageal carcinoma, and 31 gastric carcinoma samples) was used to verify the diagnostic and differential diagnostic capability of the peptidome patterns blindly. An immunoassay method was used to determine serum CEA of CRC and controls. Results. A quick classifier algorithm was used to construct the peptidome patterns for identification of CRC from controls. Two of the identified peaks at m/z 741 and 7772 were used to construct peptidome patterns, achieving an accuracy close to 100% (>CEA, P < 0.05). Furthermore, the peptidome patterns could differentiate validation group with high accuracy. Conclusions. These results suggest that the ClinProt Kit combined with mass spectrometry yields significantly higher accuracy for the diagnosis and differential diagnosis of CRC.
1. Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in males and the second in females, with over 1.2 million new cancer cases and 608,700 deaths estimated to have occurred in 2008 [1]. A majority of CRC are either locally or distantly invasive at diagnosis, restricting treatment options and reducing survival rates, whereas the 5-year survival rate is extremely favorable if detected at an early stage and successfully resected [2, 3]. Therefore, early diagnosis is of importance for CRC patient prognosis [4]. Although several screening techniques, such as colonoscopy, fecal occult blood testing (FOBT), and analysis of various serial markers are recommended, the early diagnosis rate of CRC is still comparatively low [5]. So it remains to be an urgent necessity to explore effective biomarker for diagnosis of CRC.
Proteomics, concerning comprehensive protein profile changes caused by multigene alterations, are currently considered to be the most powerful tool for global evaluation of protein expression [6]. Human serum contains thousands of proteolytically derived peptides, called peptidomes, which may provide a robust correlation of the physiological and pathological process in the entire body [7, 8]. The panels of peptidome markers might be more sensitive and specific than conventionally biomarker approaches [9]. Preliminary studies have shown that there is a great interest in the low molecular weight region, particularly peptides smaller than 20 kD, which may provide a novel means of diagnosing cancer and other diseases [8, 10, 11].
Advances in mass spectrometry (MS) now permit the display of hundreds of small- to medium-sized peptides using only microliters of serum [12, 13]. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) can detect peptides with low molecular weights at necessary sensitivity and resolution, which make it a useful technique for serum peptide profiling. Furthermore, for accurate MS analysis, the peptidomes fractionation procedure and preanalytical conditions of peptidomes mapping must be carefully assessed [14]. Magnetic bead (MB), based on nanomaterials, has been developed and was considered as a promising material for convenient and efficient enrichment of peptides and proteins in biological samples [15, 16]. Combination of MALDI-TOF MS and MS enables high throughput and sensitive investigation of peptides and proteins.
A well-defined novel technology platform, called ClinProt (Bruker Daltonics, Ettlingen, Germany), comprising a weak cationic-exchanger magnetic beads- (WCX-MB-) based sample separation, MALDI-TOF MS for peptide profiling acquisition, and a bioinformatics package for inspection and comparison of data sets to create “disease-specific” peptidome pattern models, which could serve as a powerful tool for the diagnosis of cancer [17–19].
In the current study, we used ClinProt to determine serum peptidome patterns for CRC diagnosis. The resulting spectra between groups were analysed using postprocessing software ClinProt 2.2 and patterns recognition Quick Classifier (QC) Algorithm. Diagnostic model, comprised by two differentially expressed peptides, were established and validated by the QC Algorithm, by which different groups were discriminated effectively. The diagnostic model obtained in this manner was further verified in blinded CRC, colorectal adenoma and health volunteer samples. Furthermore, to understand its differential diagnosis potential, the obtained diagnostic model was verified in blinded esophageal carcinoma (EC) and gastric cancer (GC) samples. Thus, the preliminary work was completed for an early diagnosis and differential diagnosis of CRC from an integrated perspective of peptide mass patterns.
2. Materials and Methods
2.1. Reagents and Instruments
The AutoFlex III MALDI-TOF MS, MTP 384 target plate polished steel, α-cyano-hydroxycinnamic (CHCA), MB-WCX kit and peptide calibration standard were purchased from Bruker Daltonics (Leipzig, Germany). Trifluoroacetic acid (TFA) was purchased from Alfa Aesar (Ward Hill, MA, USA). Acetonitrile (ACN) was acquired from Sigma (St. Louis, MO, USA). Diagnostic Kit of Carcinoembryonic antigen (CEA) (ELISA) was acquired from Roche Diagnostics GmbH (Sandhofer Strasse, Germany).
2.2. Patients and Sample Collection
Sixty-five health subjects (blood donor volunteers), 72 CRC patients, 16 colorectal adenoma patients, 36 EC, and 31 GC patients were enrolled with the permission of the Local Ethical Commission, and blood was collected after informed consent from the patients. Enteroscopy were performed in all health subjects to exclude the presence of incidental colon and rectum mass. Colorectal adenoma were diagnosed by enteroscopy and pathologic diagnosis. CRC, EC, and GC patients were underwent clinical, staging after surgical excision of the lesion according to the 2004 tumor-node-metastasis (TNM) stage classification [20].
Serum samples were prepared by collecting blood in a vacuum tube and allowing it to clot for 30 minutes at room temperature. About 1 mL of serum was obtained after centrifugation at 1100 g for 10 minutes and stored in small aliquots at −80°C until analysis.
2.3. Study Design
The data set including 65 controls and 72 CRC patients was randomly split into 2 groups, the clinical characteristics of CRC patients were shown in Table 1. The first group (model construction data set: 32 health volunteers and 38 CRC patients) was used for the identification of signals related to peptides expressed differentially in CRC patients compared with controls and patterns recognition. The second group (external evaluation data set: 33 health volunteers, 34 CRC patients, 16 colorectal adenoma patients, 36 EC patients and 31 GC patients) was used for the independent patterns validation of the cluster blindly. The accuracy of the peptide model was compared with that of CEA.
Table 1.
Clinical characteristics of colorectal cancer patients recruited in model construction group and external evaluation group.
| Clinical characteristics | Model construction group (n = 38) | External evaluation group (n = 34) | P value |
|---|---|---|---|
| Gender: female/male | 23/15 | 19/15 | 0.690 |
| Age (years, ) | 62.00 ± 10.95 | 61.74 ± 11.61 | 0.854 |
| TNM stage | 0.858 | ||
| I | 5 | 5 | |
| II | 18 | 15 | |
| III | 11 | 12 | |
| IV | 4 | 2 | |
| Tumor localization | 0.292 | ||
| Colon | 24 | 21 | |
| Rectum | 14 | 13 |
The gender ratio (male/female) of health volunteers, colorectal adenoma patients, EC and GC patients was 1.24, 1.67, 1.25, and 2.44, respectively. The mean age (years) of health volunteers, colorectal adenoma patients, EC, and GC patients was 54.63 ± 1.37, 59.75 ± 20.62, 61.14 ± 6.82, 58.48 ± 10.60. The difference of age and gender of health volunteers in model construction group and external evaluation data were not significant. No significant differences were either observed for age and gender between CRC and health volunteers, nor for TNM stage of CRC between model construction group and external evaluation group.
2.4. Sample Purification
We used MB-WCX for peptidome separation of samples following the standard protocol by the manufacturer [21]. Step 1, 10 μL of WCX-MB-binding solution and 10 μL of WCX-beads were combined in a 0.5 mL microfuge tube after thoroughly vortexing both reagents. Step 2, 5 μL of serum sample was added to the microfuge tube containing 10 μL of WCX-MB-binding solution and 10 μL of WCX-beads, and mixed by pipetting up and down. Step 3, microfuge tubes were then placed in a magnetic bead separator (MBS) and agitated back and forth 10 times. The beads were collected on the wall of the tubes in the MBS 1 minute later. Step 4, the supernatant was removed carefully by using a pipette. Step 5: 100 μL of WCX-MB wash buffer was added to tubes, which were agitated back and forth in the MBS 10 times. The beads were collected on the wall of the tubes, and supernatant was removed carefully by using a pipette. After three washes, 5 μL of WCX-MB elution buffer was added to disperse beads in tubes by pipetting up and down. The beads were collected on wall of tubes for 2 minutes and the clear supernatant was transferred into fresh tubes, then 5 μL WCX-MB stabilization solutions were added to the collected supernatant, mixing intensively by pipetting up and down, the mixture was then ready for spotting onto MALDI-TOF MS targets and measurement. Finally, prior to the MALDI-TOF MS analysis, we prepared targets by spotting 1 μL of the proteome fraction on the polished steel target (Bruker Daltonics ). After air drying, 1 μL of 3 mg/mL CHCA in 50% ACN and 50% Milli-Q with 2% TFA was applied onto each spot, and the target was air dried again (co-crystallization). The peptide calibration standard (1 pmol/μL peptide mixture) was applied for calibrating the machine.
2.5. Mass Spectrometry Analysis
For proteome analysis, we used a linear Autoflex III MALDI-TOF-MS with the following setting: ion source 1, 20.00 kV; ion source 2, 18.60 kV; lens, 6.60 kV; pulsed ion extraction, 120 ns; Ionization was achieved by irradiation with a crystal laser operating at 200.0 Hz. For matrix suppression, we used a high-gating factor with signal suppression up to 600 Da. Mass spectra were detected using linear positive mode. Mass calibration was performed with the calibration mixture of peptides and proteins in the mass range of 1000–12000 Da. We measured three MALDI preparations (MALDI spots) for each MB fraction. For each MALDI spot, 1600 spectra were acquired (200 laser shots at 8 different spot positions). Spectra were collected automatically using the autoflex Analysis software (Bruker Daltonik) for fuzzy controlled adjustment of critical instrument settings to generate raw data of optimized quality.
2.6. Bioinformatics and Statistical Analysis
The ClinProt Tools software 2.2 (Bruker Daltonik) was used for analysis of all serum sample data derived from either patients or normal controls. Data analysis began with raw data pretreatment, including baseline subtraction of spectra, normalization of a set of spectra, internal peak alignment using prominent peaks, and a peak picking procedure. The pretreated data were then used for visualization and statistical analysis in ClinProt Tools. Statistically significant different quantity of peptides was determined by means of Welch's t-tests. The significance was set at P < 0.05. Class prediction model was set up by QC Algorithm. A classify peptidome patterns was constructed. To determine the accuracy of the class prediction, firstly, a cross-validation was implemented. Twenty percent of model construction group were randomly selected sample as a test set, and the rest samples were taken as a training set in the class predictor algorithm. Secondly, designed as double blind test, the samples of external evaluation group were classified by the classify peptidome patterns constructed by QC Algorithm.
2.7. Detection of CEA
The serum CEA of 38 CRC and 32 health volunteers included in model construction group was detected using an electrochemiluminescent immunoassay method following the standard protocol by the manufacturer (The methods were omitted). The sample was diagnosed as CRC (CEA⩾5 ng/mL), otherwise diagnosed as health volunteers (CEA < 5 ng/mL).
2.8. Statistical Methods, Evaluation of Assay Precision
We analyzed each spectrum obtained from MALDI-TOF MS with Autoflex analysis and ClinProt TM software (Bruker Daltonics), the former to detect the peak intensities of interest and the latter to compile the peaks across the spectra obtained from all samples. This allowed differentiation between the cancer and control samples. To evaluate the precision of the assay, we determined within- and between-run variations by use of multiple analyses of bead fractionation and MS for 2 plasma samples. For within- and between-run variation, we examined 3 peaks with various intensities. We determined within-run imprecision by evaluating the CVs for each sample, using 8 assays within a run, then determined between-run imprecision by performing 8 different assays over a period of 7 days. SPSS 16.0 was used for analysis of the clinical characteristics of volunteers using χ2 test or t-test. The significance was set at P < 0.05. Also, SPSS 16.0 was used to compare the accuracy of the peptidome models and CEA.
3. Results
For the reproducibility of the protein profiling, Within- and between-run reproducibility of 2 samples were determined with the WCX-MB fractionation and MALDI-TOF MS analysis. In each profile, 3 peaks with different molecular masses were selected to evaluate the precision of the assay. Despite varying peptide masses and spectrum intensities, the peak CVs were all <3% in the within-run and <9% in the between-run assays. These values were consistent with the reproducibility data for the Protein Biology System reported by the manufacturer (Bruker Daltonik).
In the pilot study we evaluated the differences of the serum proteome profiles of CRC in comparison to health subjects. The mass spectra from 1 to 18 kDa were obtained by MALDI-TOF MS in linear mode. The representative mass spectra of prefractionated serum of model construction group are reported in Figure 1. On average about 156 signals common to the two groups have been detected in this mass range and about 61 were identified by the ClinProt software with a statistically different area (P < 0.05 by Wilcoxon analysis) in model construction population, including 49 upregulated and 12 downregulated peptides. Two peptides selected for model construction were shown in Table 2 and the others are shown in Supplementary Table S1 (see Supplementary Material available online at doi;10.1155/2012/985020).
Figure 1.
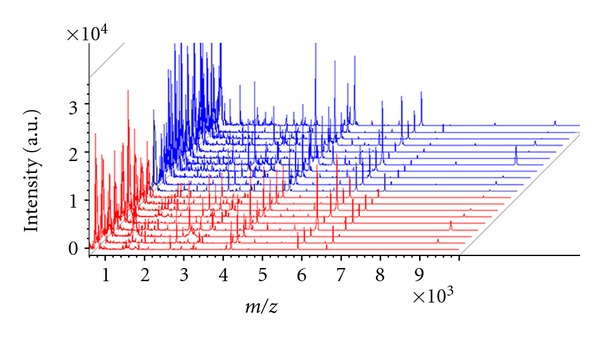
View of the aligned mass spectra of the serum protein profile of model construction group (red: 10 health subjects, blue: 10 colorectal cancer patients) obtained by MALDI-TOF after purification with WCX magnetic beads.
Table 2.
Statistic of the 2 candidate biomarkers signals selected for the diagnostic model for identifying colorectal cancer from health individuals.
| Mass | PW1 | PAD2 | Ave(C)3 | Ave(N)4 | SD(C)5 | SD(N)6 |
|---|---|---|---|---|---|---|
| 7772.38 | <0.000001 | <0.000001 | 232.5 | 6.75 | 111.94 | 10.39 |
| 741.43 | <0.000001 | <0.000001 | 43.32 | 214.97 | 32.09 | 57.61 |
1 P value calculated with the Wilcoxon test; values lower than 0.05 suggest statistical relevance.
2 P value calculated with the Anderson-Darling test, values lower than 0.05 suggest statistical relevance.
3Average area of peaks for colorectal cancer subjects.
4Average area of peaks for health subjects.
5Standard deviation of peaks for colorectal cancer subjects.
6Standard deviation of peaks for health subjects.
Classification models were developed to classify samples between CRC and health volunteers. The use of individual peaks as diagnostic biomarker for CRC was addressed using QC algorithm analysis. First, we conducted comparison between CRC and health volunteers. Second, all detected peaks were analysed by ClinProt 2.2 to generate cross-validated classification models. The optimized model resulted in the following correct classification of samples. Two peptide ion signatures (m/z 741 and 7772) was provided as a class prediction for a cross-validation set to discriminate CRC from health volunteers, which achieved a recognition capacity of 97.3% and a cross-validation of 97.3%. Regions of the mass spectra obtained at 800 resolving are reported in Figure 2.
Figure 2.
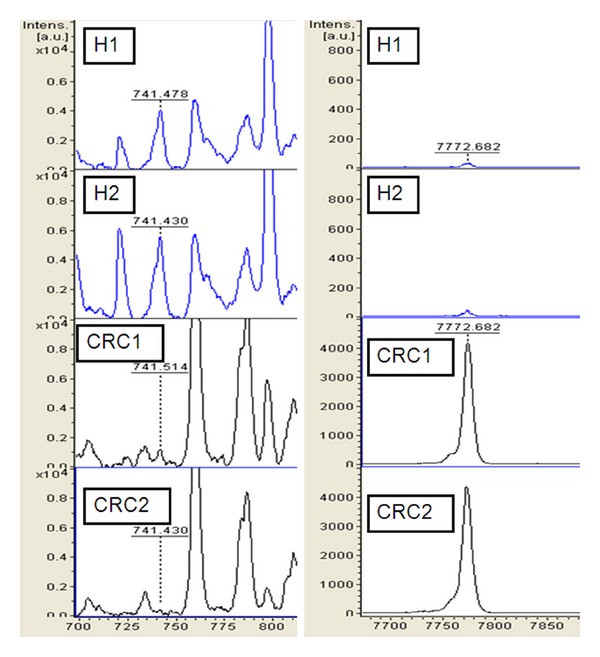
Zoom of the mass range for the two signals (MALDI-TOF linear mode) used in the cluster to differentiate colorectal cancer (CRC) from healthy volunteers (H).
Preliminary statistical analysis was carried out for each single marker and for the cluster of signals by the receiver operating characteristic curve analysis. Area under curve (AUC) of peak A at m/z 741 (P < 0.000001) and of peak B at m/z 7442 (P < 0.000001) was 0.988 and 0.991, respectively, which corresponds to a highly accurate test, according to the criteria suggested by Swets [22] (Figure 3). Moreover areas of these peaks in the spectra of CRC were statistically different from those of the health volunteers (Figure 4). Combination of the two peaks allowed to yielding a specificity of 100%, and a sensitivity of 94.74% for CRC (Table 3, Figure 5).
Figure 3.
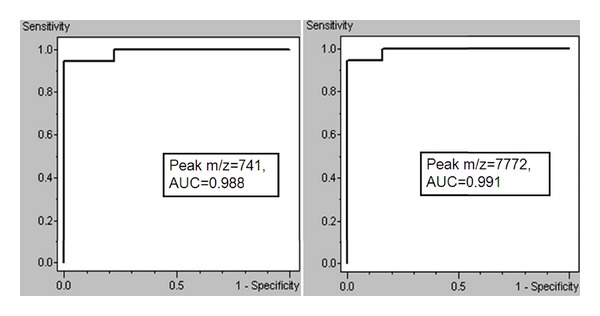
The receiver operating characteristic curve of two signals selected for the diagnostic model. AUC, areas under the receiver operating characteristic curve.
Figure 4.
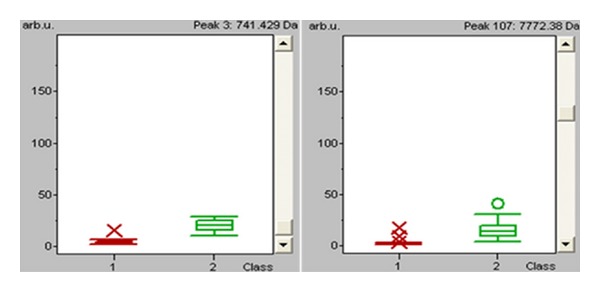
Box-and-whiskers plot calculated from the areas of the two signals used in the cluster for the two studied populations. Red represents colorectal cancer, green represents healthy volunteers.
Table 3.
The predicted results of peptidome pattern distinguishing colorectal cancer patients from controls.
| Group | Colorectal cancer | Controls | Sensitivity (%) | Specificity (%) | Youden's index | |||
|---|---|---|---|---|---|---|---|---|
| Health | Colorectal adenoma | Esophageal carcinoma | Gastric cancer | |||||
| Model construction | 38 | 32 | — | — | — | 94.74% (36/38) | 100% (32/32) | 0.95 |
| External validation | ||||||||
| I | 34 | 33 | — | — | — | 94.12% (32/34) | 100% (33/33) | 0.94 |
| II | 34 | — | 16 | — | — | 94.12% (32/34) | 100% (16/16) | 0.94 |
| III | 34 | — | — | 36 | — | 94.12% (32/34) | 100% (36/36) | 0.94 |
| IV | 34 | — | — | — | 31 | 94.12% (32/34) | 100% (31/31) | 0.94 |
Figure 5.
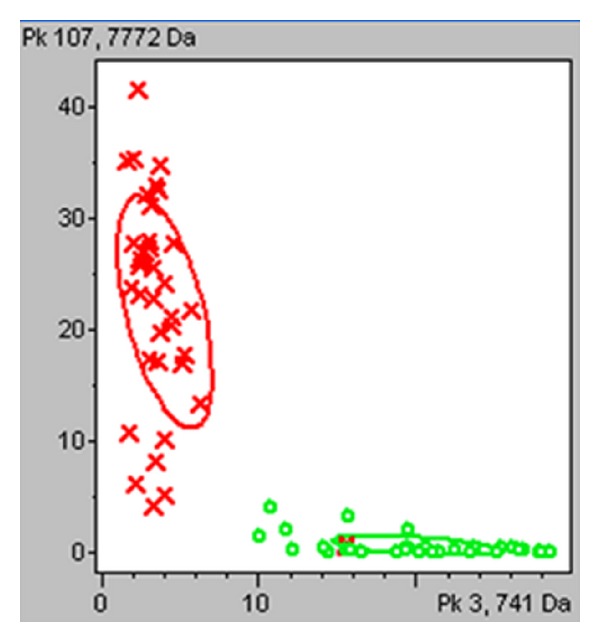
Two-dimensional peak distribution view of the two peaks selected for the diagnostic model. The peak area and the m/z values are indicated on the x- and y-axes. The ellipses represent the standard deviation of the class average of the peak areas/intensities. Red represents colorectal cancer patients and green represents healthy volunteers.
To verify the accuracy of the established QC classification model with the adopted peptides, we introduced another group of samples (not used in model construction), which consisted of 34 CRC, 16 colorectal adenoma, and 33 health volunteers. As a result, the model correctly classified 94.12% (32/34) of CRC (sensitivity), 100% (16/16) of colorectal adenoma (specificity), and 100% (33/33) of health volunteers (specificity), which surpassed that of CEA (a specificity of 51.02% (25/49), and a sensitivity of 41.18% (14/34)). To verify the differential diagnosis ability of the QC classification model, we introduced a group of other common cancers samples, which consisted of 36 EC and 31 GC. As a result, the model correctly classified 100% (36/36) of EC (specificity) and 100% (31/31) GC (specificity) as controls (Table 3).
4. Discussion
The usefulness of multiple markers for diagnosis, prognosis, and for predicting the risk of developing diseases or their complications is now widely recognized [7, 23]. Various proteomic approaches have been applied to biomarker discovery using biological fluids. It is being interestingly recognized that low mass weight peptides, such as S100A8 and fibrinogen, play an important role in physiological and pathological process and could be used as relevant biomarker candidates [24, 25]. Recently mass spectrum that directly detects and differentiates short peptides has offered a promising approach for peptidomic biomarker discovery [8, 10, 26–28].
Compared with genomic approaches, proteomic analysis has the advantage of visualizing co- and posttranslational modifications of proteins, possibly of relevance for biologic function. Alternative approaches for measuring polypeptides are time-consuming for routine use, such as the classic method of comparing data from two-dimensional electrophoresis, subsequent isolation of the proteins from the gel, and analysis by MS [29]. Another method, the surface-enhanced laser desorption and ionization time-of-flight MS, recently reported by several groups, have been applied in common cancers screening using serum peptidome patterns [30–33]. These reports emphasized the potential diagnosis value of low molecular mass peptide or protein.
MALDI is a soft ionization technique used in MS, allowing the analysis of bio-molecules such as proteins, peptides sugars, and large organic molecules. The time-of-flight (TOF) mass spectrometer is ideally suited type to the MALDI, which can reach a resolving power m/Δm of the well above 20,000 FWHM (full-width half-maximum; Δm defined as the peak width at 50% of peak height). As a powerful tool for surveying complex patterns of biologically informative molecules, MALDI-TOF MS protein profiling has been applied in proteomics biomarker research and has become a promising tool in cancer biomarker research [26, 34, 35].
In present study, by integrating the purification of short peptides with WCX-MB, detection of peak intensity with MALDI-TOF MS, and profile analysis with ClinProt Tool software 2.2, we have successfully detected a series of short peptides that differentially expressed in the serum of patients with CRC. A case control comparative analysis between CRC and health volunteer was performed. Peptidomic maps associated with the disease were drawn. The results show that, compared to normal controls, CRC sharing 61 significantly differentiated peptides, including 49 upregulated and 12 downregulated peptides. Current knowledge of cellular regulation indicates that many networks operate at the epigenetic, transcriptional, and translational levels. Genomic and proteomic technologies will help further understand the intracellular signaling and gene transcription systems as well as the protein pathways that connect extracellular microenvironment to the serum or plasma macroenvironment of cancer [36]. These 61 interesting significantly differentiated peptides may provide further evidence for understanding the occurrence and progress of CRC. In particular, the prominent peptides that have a greater than twofold change in intensity, such as m/z 741, 7772, 5907, may be defined as the leading differential peptides associated with colorectal cancer, worthy of further sequence determination and function analysis.
By using the QC algorithm analysis, classification model were developed to classify samples between healthy volunteers and CRC. A cluster of two peptides at m/z 741 and 7772 achieved a recognition capacity and a cross-validation of close to 100% (a specificity of 100%, and a sensitivity of 94.74%) to discriminate CRC from healthy volunteers. Blinded verification of the QC classification model proved to correctly classify 94.12% (32/34) of CRC, 100% (33/33) health volunteers. Furthermore, to evaluate the differential diagnosis capacity, 16 colorectal adenoma patients and 36 EC patients, and 31 GC patients were applied for blinded verification. Interestingly, 100% of the individuals were classified as control, which suggest that the classification model could identify CRC from colorectal adenoma and two of the most common digestive tract cancers (EC and GC). This demonstrated that the QC Algorithm would be effective in facilitating the construction of a sensitive and specific diagnostic model.
According to our knowledge, this study is the first to screen CRC related short peptides in sera by combining WCX-MB and MALDI-TOF-MS. The classification model we have setup have application in providing alternatives for CRC diagnosis or differential diagnosis, and may provide a better understanding of the pathogenesis in CRC or help in tailoring the use of chemotherapy to each patient, finally resulting in an improvement in patient outcome. Despite of the high sensitivity and specificity, the number of specimens analyzed in this study was relatively small, which may limit the validity of the results. The next step of our study will be to analyse larger patient cohorts and to run blinded samples to confirm the usefulness of our currently identified peptides for CRC diagnosis. After this confirmation, we will then isolate and identify the biomarkers of the interest and study their biological role in CRC pathogenesis.
In conclusion, we directly profiled peptidome patterns from WCX-MB-purified serum samples with MALDI-TOF MS, and constructed a peptidome model that differentiated CRC from control samples with high sensitivity and specificity, which may be applied as an alternative method for the diagnosis and differential diagnosis of CRC.
Supplementary Material
Table 1: Statistic of the 61 candidate biomarkers for identifying colorectal cancer patients from healthy volunteers.
Conflict of Interests
The authors stated that there are no conflict of interests regarding the publication of this paper.
Acknowledgment
This work was funded by the Medical Science Research Project of The Chinese People's Liberation Army (no. 08Z006).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer Journal for Clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer Journal for Clinicians. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Ciccolallo L, Capocaccia R, Coleman MP, et al. Survival differences between European and US patients with colorectal cancer: role of stage at diagnosis and surgery. Gut. 2005;54(2):268–273. doi: 10.1136/gut.2004.044214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards BK, Ward E, Kohler BA, et al. Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010;116(3):544–573. doi: 10.1002/cncr.24760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith RA, Cokkinides V, Brooks D, Saslow D, Shah M, Brawley OW. Cancer screening in the United States, 2011: a review of current American Cancer Society guidelines and issues in cancer screening. CA Cancer Journal for Clinicians. 2011;61(1):8–30. doi: 10.3322/caac.20096. [DOI] [PubMed] [Google Scholar]
- 6.Phizicky E, Bastiaens PIH, Zhu H, Snyder M, Fields S. Protein analysis on a proteomic scale. Nature. 2003;422(6928):208–215. doi: 10.1038/nature01512. [DOI] [PubMed] [Google Scholar]
- 7.Issaq HJ, Xiao Z, Veenstra TD. Serum and plasma proteomics. Chemical Reviews. 2007;107(8):3601–3620. doi: 10.1021/cr068287r. [DOI] [PubMed] [Google Scholar]
- 8.Dai Y, Hu C, Wang L, et al. Serum peptidome patterns of human systemic lupus erythematosus based on magnetic bead separation and MALDI-TOF mass spectrometry analysis. Scandinavian Journal of Rheumatology. 2010;39(3):240–246. doi: 10.3109/03009740903456292. [DOI] [PubMed] [Google Scholar]
- 9.Petricoin EF, Belluco C, Araujo RP, Liotta LA. The blood peptidome: a higher dimension of information content for cancer biomarker discovery. Nature Reviews Cancer. 2006;6(12):961–967. doi: 10.1038/nrc2011. [DOI] [PubMed] [Google Scholar]
- 10.Liu LH, Shan BE, Tian ZQ, et al. Potential biomarkers for esophageal carcinoma detected by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clinical Chemistry and Laboratory Medicine. 2010;48(6):855–861. doi: 10.1515/CCLM.2010.138. [DOI] [PubMed] [Google Scholar]
- 11.Sui W, Huang L, Dai Y, Chen J, Yan Q, Huang H. Proteomic profiling of renal allograft rejection in serum using magnetic bead-based sample fractionation and MALDI-TOF MS. Clinical and Experimental Medicine. 2010;10(4):259–268. doi: 10.1007/s10238-010-0094-5. [DOI] [PubMed] [Google Scholar]
- 12.Koomen JM, Li D, Xiao LC, et al. Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. Journal of Proteome Research. 2005;4(3):972–981. doi: 10.1021/pr050046x. [DOI] [PubMed] [Google Scholar]
- 13.Villanueva J, Philip J, Entenberg D, et al. Serum peptide profiling by magnetic particle-assisted, automated sample processing and MALDI-TOF mass spectrometry. Analytical Chemistry. 2004;76(6):1560–1570. doi: 10.1021/ac0352171. [DOI] [PubMed] [Google Scholar]
- 14.Omenn GS. The Human Proteome Organization Plasma Proteome Project pilot phase: reference specimens, technology platform comparisons, and standardized data submissions and analyses. Proteomics. 2004;4(5):1235–1240. doi: 10.1002/pmic.200300686. [DOI] [PubMed] [Google Scholar]
- 15.Yao N, Chen H, Lin H, Deng C, Zhang X. Enrichment of peptides in serum by C8-functionalized magnetic nanoparticles for direct matrix-assisted laser desorption/ionization time-of-flight mass spectrometry analysis. Journal of Chromatography A. 2008;1185(1):93–101. doi: 10.1016/j.chroma.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 16.Whiteaker JR, Zhao L, Zhang HY, et al. Antibody-based enrichment of peptides on magnetic beads for mass-spectrometry-based quantification of serum biomarkers. Analytical Biochemistry. 2007;362(1):44–54. doi: 10.1016/j.ab.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.West-Nørager M, Kelstrup CD, Schou C, Høgdall EV, Høgdall CK, Heegaard NHH. Unravelling in vitro variables of major importance for the outcome of mass spectrometry-based serum proteomics. Journal of Chromatography B. 2007;847(1):30–37. doi: 10.1016/j.jchromb.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 18.Shin S, Cazares L, Schneider H, et al. Serum biomarkers to differentiate benign and malignant mammographic lesions. Journal of the American College of Surgeons. 2007;204(5):1065–1071. doi: 10.1016/j.jamcollsurg.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 19.Baumann S, Ceglarek U, Fiedler GM, Lembcke J, Leichtle A, Thiery J. Standardized approach to proteome profiling of human serum based on magnetic bead separation and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clinical Chemistry. 2005;51(6):973–980. doi: 10.1373/clinchem.2004.047308. [DOI] [PubMed] [Google Scholar]
- 20.Compton CC, Greene FL. The staging of colorectal cancer: 2004 and beyond. Ca-A Cancer Journal for Clinicians. 2004;54(6):295–308. doi: 10.3322/canjclin.54.6.295. [DOI] [PubMed] [Google Scholar]
- 21.Ketterlinus R, Hsieh SY, Teng SH, Lee H, Pusch W. Fishing for biomarkers: analyzing mass spectrometry data with the new ClinProTools software. BioTechniques. 2005;(supplement):37–40. doi: 10.2144/05386su07. [DOI] [PubMed] [Google Scholar]
- 22.Swets JA. Measuring the accuracy of diagnostic systems. Science. 1988;240(4857):1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- 23.Zethelius B, Berglund L, Sundström J, et al. Use of multiple biomarkers to improve the prediction of death from cardiovascular causes. The New England Journal of Medicine. 2008;358(20):2107–2116. doi: 10.1056/NEJMoa0707064. [DOI] [PubMed] [Google Scholar]
- 24.Tolson JP, Flad T, Gnau V, et al. Differential detection of S100A8 in transitional cell carcinoma of the bladder by pair wise tissue proteomic and immunohistochemical analysis. Proteomics. 2006;6(2):697–708. doi: 10.1002/pmic.200500033. [DOI] [PubMed] [Google Scholar]
- 25.Cheng AJ, Chen LC, Chien KY, et al. Oral cancer plasma tumor marker identified with bead-based affinity-fractionated proteomic technology. Clinical Chemistry. 2005;51(12):2236–2244. doi: 10.1373/clinchem.2005.052324. [DOI] [PubMed] [Google Scholar]
- 26.Alagaratnam S, Mertens BJA, Dalebout JC, et al. Serum protein profiling in mice: identification of Factor XIIIa as a potential biomarker for muscular dystrophy. Proteomics. 2008;8(8):1552–1563. doi: 10.1002/pmic.200700857. [DOI] [PubMed] [Google Scholar]
- 27.Chinello C, Gianazza E, Zoppis I, et al. Serum biomarkers of renal cell carcinoma assessed using a protein profiling approach based on ClinProt technique. Urology. 2010;75(4):842–847. doi: 10.1016/j.urology.2009.09.050. [DOI] [PubMed] [Google Scholar]
- 28.Du J, Yang S, Lin X, et al. Use of anchorchip-time-of-flight spectrometry technology to screen tumor biomarker proteins in serum for small cell lung cancer. Diagnostic Pathology. 2010;5, article 60 doi: 10.1186/1746-1596-5-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ikonomou G, Samiotaki M, Panayotou G. Proteomic methodologies and their application in colorectal cancer research. Critical Reviews in Clinical Laboratory Sciences. 2009;46(5-6):319–342. doi: 10.3109/10408360903375277. [DOI] [PubMed] [Google Scholar]
- 30.Petricoin EF, Ardekani AM, Hitt BA, et al. Use of proteomic patterns in serum to identify ovarian cancer. The Lancet. 2002;359(9306):572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 31.Chen YD, Zheng S, Yu JK, Hu X. Artificial neural networks analysis of surface-enhanced laser desorption/ionization mass spectra of serum protein pattern distinguishes colorectal cancer from healthy population. Clinical Cancer Research. 2004;10(24):8380–8385. doi: 10.1158/1078-0432.CCR-1162-03. [DOI] [PubMed] [Google Scholar]
- 32.Liu XP, Shen J, Li ZF, Yan L, Gu J. A serum proteomic pattern for the detection of colorectal adenocarcinoma using surface enhanced laser desorption and ionization mass spectrometry. Cancer Investigation. 2006;24(8):747–753. doi: 10.1080/07357900601063873. [DOI] [PubMed] [Google Scholar]
- 33.Fan N-J, Gao C-F, Wang X-L. Identification of regional lymph node involvement of colorectal cancer by serum SELDI proteomic patterns. Gastroenterology Research and Practice. 2011;2011:6 pages. doi: 10.1155/2011/784967.784967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hortin GL. The MALDI-TOF mass spectrometric view of the plasma proteome and peptidome. Clinical Chemistry. 2006;52(7):1223–1237. doi: 10.1373/clinchem.2006.069252. [DOI] [PubMed] [Google Scholar]
- 35.Navare A, Zhou M, McDonald J, Noriega FG, Sullards MC, Fernandez FM. Serum biomarker profiling by solid-phase extraction with particle-embedded micro tips and matrix-assisted laser desorption/ionization mass spectrometry. Rapid Communications in Mass Spectrometry. 2008;22(7):997–1008. doi: 10.1002/rcm.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho WCS. Contribution of oncoproteomics to cancer biomarker discovery. Molecular Cancer. 2007;6, article 25 doi: 10.1186/1476-4598-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1: Statistic of the 61 candidate biomarkers for identifying colorectal cancer patients from healthy volunteers.


