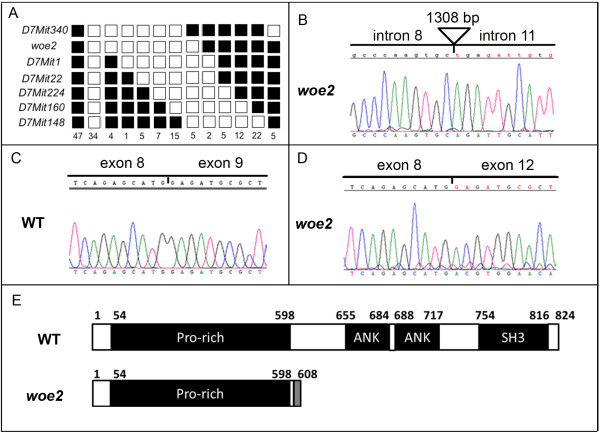
Figure 4.
The genetic analysis of the woe2 locus. The woe2 locus was mapped to mouse chromosome 7 (A) between D7Mit340 and D7Mit1 establishing Ppp1r13l as a candidate gene. Each column represents the haplotype found in progeny of woe2 mutants (C57BLX129/SvJ background) when backcrossed to the C3A.BliAPde6+/J strain. Black boxes represent the C3ABliA-Pde6+/J allele and white boxes represent the C57BL6X129/SvJ allele. The number of offspring inheriting each haplotype is listed at the bottom of each column. Sequence analysis of genomic DNA from woe2 tissues (B) identified a 1308 bp deletion encompassing the genomic region from Ppp1r13l intron 8 to intron 11. RT-PCR analysis from WT tissues (C) identified a single Ppp1r13l transcript that matched the Ppp1r13l reference sequence (NM_001010836). RT-PCR analysis from woe2 tissues (D) identified a single aberrantly spliced Ppp1r13l transcript with missing exons 9, 10 and 11. (E) depicts a schematic of wild type PPP1R13L protein (NP_001010836) (top) conserved domains: Pro-rich domain (54–598), ANK1 domain (655–684), ANK2 (688–717), and SH3 (754–816) domain. The woe2 putative PPP1R13Lp.E602TX7 protein (bottom figure) shows, following Met601, an addition of 7 novel amino acids (TWNKAWD) depicted by a gray box, a premature stop and a loss of 223 amino acids from the C-terminus encompassing ANK1, ANK2 and SH3 domains. Numbers on the top of each figure in (E) represent the amino acid residues.