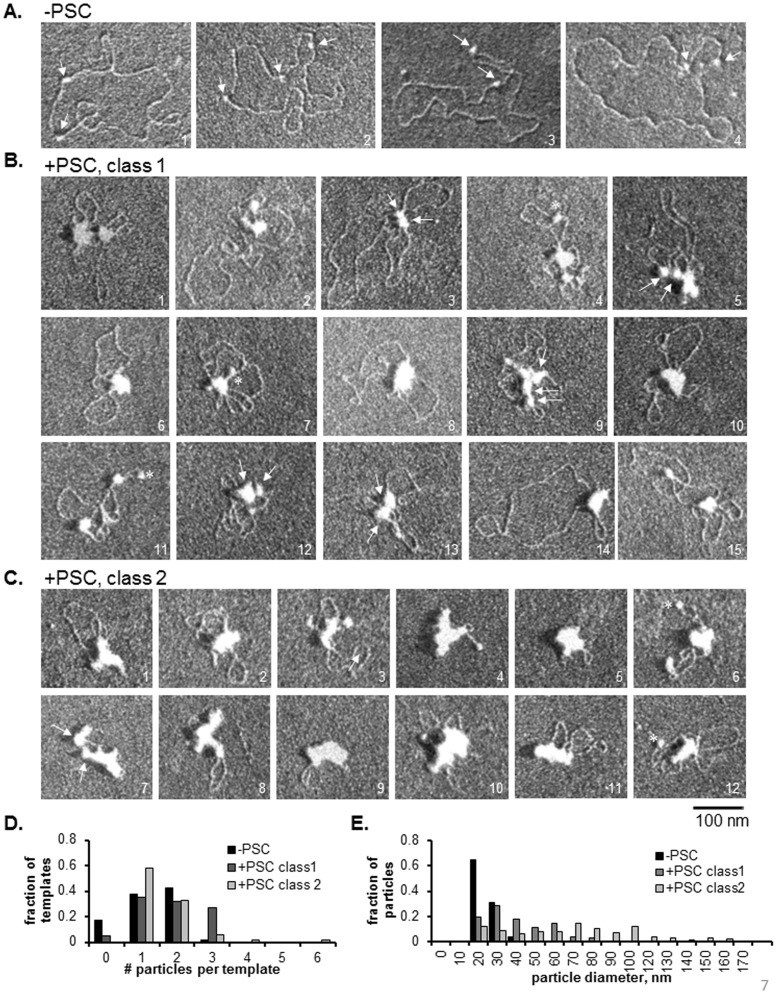
Figure 7. PSC clusters nucleosomes and DNA on sparsely assembled plasmids.
(a) Representative EM images of plasmids with two 601 nucleosome positioning sequences assembled at low ratio of histones to DNA. Note that fully assembled plasmids would contain 17 nucleosomes and 601 sequences are separated by 385 base pairs. Plasmids were assembled in the presence of E.coli Topoisomerse I so that plasmids are relaxed. Arrows point to nucleosomes. Note that template 2 was the only observed example of more than 2 nucleosomes on the plasmid (3), out of the 103 molecules that were analyzed (Table 1). (b) Sparsely assembled plasmids with PSC. Class 1 molecules (see text) are more extended, and likely have fewer copies of PSC bound than Class 2 molecules (c), which are highly compacted. Arrows point to particles (likely to be PSC bound nucleosomes) that have come together and may represent the bridged configuration. Note that in some cases, more than one template may be clustered (such as molecule 3). Asterisks indicate particles that may be unbound nucleosomes (based on their size), although they could also be bound PSC. (c) Sparsely assembled plasmids with PSC with Class 2 configurations. Note that the molecules represent a series between the most extended Class 1 molecules and the most highly compacted Class 2 molecules. (d) Summary of the number of particles per template. The finding that Class 1 molecules frequently have more than 2 particles indicates that PSC must bind to naked DNA (as well as nucleosomes) on some templates. See Table 1 for summary of measurements from this and a similar experiment.