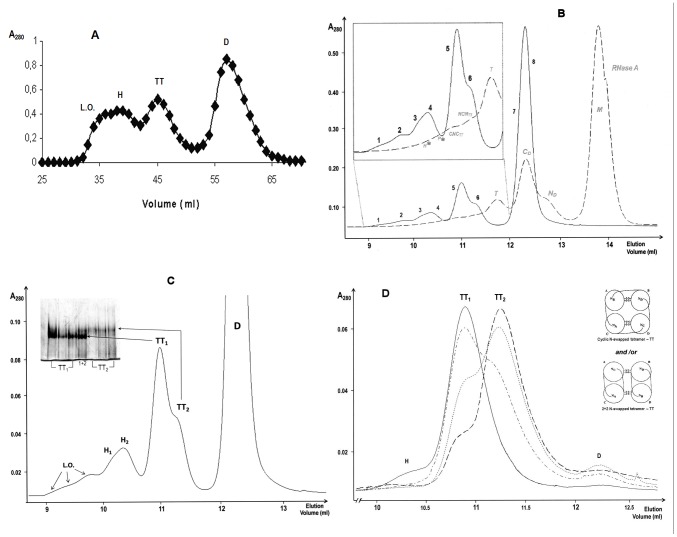
Figure 1. SEC chromatograms and PAGE under non denaturing conditions of BS-RNase aggregates obtained by lyophilising the protein from a 40% (v/v) acetic acid solution.
(A) SEC pattern obtained with a Sephadex G100 column. Elution with ammonium acetate 0.1 M, pH 5.65, flow rate of 0.4 ml/min. (B) SEC chromatogram of BS-RNase multimers superimposed with that of RNase A oligomers: both patterns were obtained with a Superdex 75 10/300 GL column. Elution with 0.2 M NaPi, pH 6.7, flow rate 0.1 ml/min. (C) Enlarged Superdex 75 SEC pattern of BS-RNase aggregates; in the inset, 7.5% non denaturing PAGE of the two BS-tetramers, run-time 110 min. (D) Additional purification of the two BS-RNase tetramers: their mixture was concentrated to 25 µl in 0.4 M NaPi, and re-chromatographed in the Superdex 75 column equilibrated with the same buffer (dashed+dotted line). Then, TT1 and TT2 fractions were further purified: once for TT1, continuous line; twice for TT2, dotted and dashed lines, respectively. In the right part of the panel are reported the models of two N-swapped BS-RNase tetramers proposed by Adinolfi et al. [13]: they cannot be associated to both tetramers. The various BS-RNase species are: D, native dimer; TT1 and TT2, two tetrameric conformers, H (1 and 2), hexamers; L.O., larger oligomers. Concerning RNase A, grey italics labels: M, native monomer, ND, N-terminal-swapped dimer, CD, C-terminal-swapped dimer; T, trimers; NCNTT: double N+C-swapped tetramer; CNCTT: double C+N-swapped tetramer; P*: pentamers; H*: hexamers. The asterisk* is present to mention that P and H positions are derived from data obtained in [21].