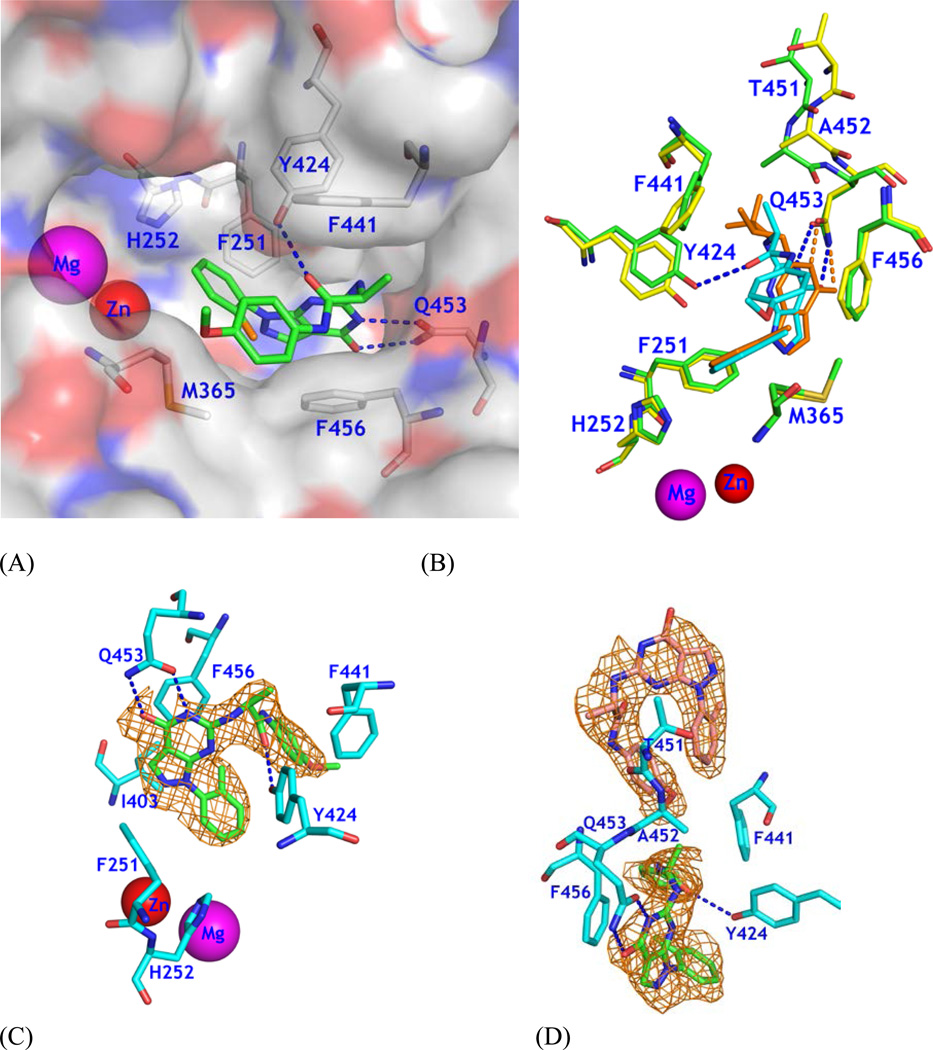
Figure 2.
Binding of 28 to PDE9A2. (A) Surface presentation on binding of inhibitor 28 (green sticks) to the active site of PDE9. The chlorine atom of 28 is in orange color. Dotted lines represent the hydrogen bonds between 28 and PDE9 residues Tyr424 and Gln453. (B) Superposition of PDE9-28 (green and cyan) over PDE9-10r (yellow and salmon). Thr451 and Ala452 show significant positional differences between PDE9-28 and PDE9-10r. The pyrazolopyrimidinone ring has about 0.7 Å difference between two structures. (C) Electron density for 28 bound to chain A of the PDE9 dimer. The mesh (Fo - Fc) map was drawn from the PDE9 structure with omission of 28 and contoured at 2.5σ. (D) Density for two molecules of 28 bound to chain B. The lower 28 binds to the active site of PDE9.