Figure 1.
RNase E Cleavage Can Be Guided by an Artificial sRNA
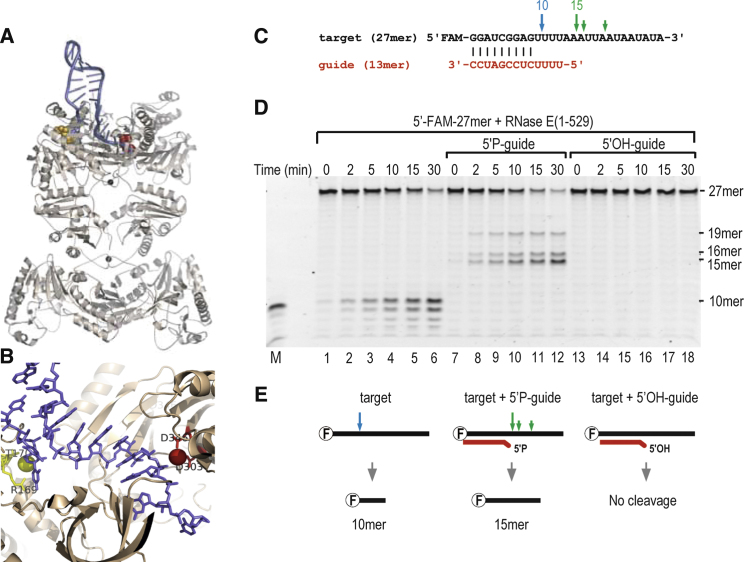
(A) Hypothetical model of the complex of the tetrameric catalytic domain of RNase E with a structured substrate, constructed from experimental crystal lattice interactions (Callaghan et al., 2005).
(B) Expanded view that includes the 5′ sensor region and the catalytic site. The spacing from the end of the seed matching duplex region to the catalytic metal is 5 to 6 bases. The RNA substrate is colored blue; in yellow are the critical residues of the 5′ sensing pocket and 5′ phosphate atom, shown as a sphere; and in red are the active site aspartate residues and the catalytic magnesium ion (sphere).
(C) The sequences of the 27-mer target RNA with a 5′ fluorescent group (FAM, black) and its partially complementary 13-mer guide (red). The guide has either a 5′ hydroxyl or a monophosphate group.
(D) Denaturing gel showing the degradation products of 27-mer alone (left panel), in presence of 5′-P-13-mer guide RNA (middle panel) and in presence of 5′-OH-13-mer guide RNA (right panel). The visualization is for the FAM group so only the 5′end degradation products are visible.
(E) Schematic of the cleavage patterns observed in (D). These are also indicated by arrows in (C) for the target 27-mer RNA in the presence (green) and absence (blue) of 13-mer guide RNA. (See also Figure S1.)