Abstract
Mutations in the lipase member H (LIPH) gene cause autosomal recessive hypotrichosis with woolly hair. We report herein on five consanguineous families from Pakistan segregating hypotrichosis and woolly hair. Genetic investigation using polymorphic microsatellite markers revealed homozygosity for a region spanning the HYPT7 locus on chromosome 3 in affected individuals of all five families. Sequence analysis of the LIPH gene revealed a novel nonsense mutation (p.Arg260X) associated with hypotrichosis without woolly hair in one family. In the remaining four families we identified previously described mutations in a homozygous state in affected members. These findings extend the spectrum of known LIPH mutations in the Pakistani population.
Autosomal recessive hypotrichosis with woolly hair (HYPT7; MIM#604379) is a rare form of alopecia characterized by sparse woolly scalp hair, sparse to absent eyebrows, eyelashes and body hair. Three genetically distinct forms of localized autosomal recessive hypotrichosis (LAH1-3) have been identified. The three forms of hypotrichosis are clinically similar and are caused by mutations mapped to chromosomes 18q12.1, 3q27.3 and 13q14.11-q21.32, respectively1,2,3. HYPT7 (LAH2) is associated with mutations in the lipase member H (LIPH) gene on chromosome 34. The enzyme lipase H (LIPH) synthesizes lysophosphatidic acid (LPA) which regulates hair follicle formation mediated by TGFα release and EGFR transactivation through the receptor P2RY55. Here we identified five families segregating autosomal recessive hypotrichosis showing autozygosity of the HYPT7 loci on chromosome 3q. Subsequent analysis of the LIPH gene revealed homozygosity for a novel truncating mutation, as well as three previously identified mutations in affected individuals.
Results
Linkage and autozygosity analysis of the five families revealed homozygosity for markers flanking the LIPH locus in affected members of all families. We then performed sequence analysis of the LIPH gene and we identified mutations segregating hypotrichosis in all five families. Affected individuals of family 1 were found homozygous for novel single nucleotide substitution at position 778 (c.778A>T) resulting in a nonsense mutation at amino acid 260 (p.Arg260X). Affected members of the other four families were shown to be homozygous for previously identified LIPH mutations. Family 2 segregates the missense mutation c.322T>C (p.Trp108Arg), Families 3 and 4 segregate the two base pair deletion c.659_660delTA (p.Ile220ArgfsX29), and Family 5 segregates the duplication c.280_369dup (p.Gly94_Lys123dup) (Table 1). The closest flanking polymorphic microsatellite markers showed different haplotypes associated with the c.659_660delTA mutation in families 3 and 4, suggesting recurrent events for the p.Ile220ArgfsX29 mutation. Each mutation segregates with the disease and parents to affected individuals who were investigated were shown to be heterozygous.
Table 1. Mutations and main phenotype in the LIPH gene in the five families.
| Family | Patients (n) | DNA | Protein | Hypotrichosis | Woolly hair | Pigmentation |
|---|---|---|---|---|---|---|
| 1 | 2 | c.778A>T | p.Arg260X | moderate | no | no |
| 2 | 8 | c.322T>C | p.Trp108Arg | moderate | no | yes |
| 3 | 4 | c.659_660delTA | p.Ile220ArgfsX29 | severe | no | no |
| 4 | 3 | c.659_660delTA | p.Ile220ArgfsX29 | mild | yes | no |
| 5 | 4 | c.280_369dup | p.Gly94_Lys123dup | mild | yes | yes |
Discussion
The novel LIPH nonsense mutation (p.Arg260X) in family 1 is predicted to result in nonsense mediated decay or a truncated LIPH protein. The mutation is associated with hypotrichosis without woolly hair in all affected members of the family. In family 3 and 4 we identified a previously identified and apparently recurrent two base pair deletion (c.659_660delTA) predicted to result in a frame shift and a degraded mRNA or a truncated protein (p.Ile220ArgfsX29). Thus, the effect of these two truncating mutations can be predicted to be similar and severe on LIPH function. This is supported by previous studies of LIPH mutations using a biochemical in vitro assay that shows a complete loss of LIPH function when introducing deleterious mutations6. Interestingly, the c.659_660delTA mutation is associated with severe hypotrichosis without woolly hair in family 3 whereas the same mutation causes mild hypotrichosis with woolly hair in family 4. In the remaining two families we identified a missense mutation associated with moderate hypotrichosis without woolly hair (family 2) and a duplication segregating mild hypotrichosis and woolly hair (family 5). The latter two mutations were previously identified in Pakistani families with autosomal recessive hypotrichosis and woolly hair7,8,9. Our findings support a considerable clinical variability associated with LIPH mutations and we were unable to detect any genotype-phenotype correlations in the patients. The LPA-mediated signalling through the receptor P2RY5 is complex and includes many interacting pathways, including the transactivation of the epidermal growth factor receptor5. This could possibly, at least in part, be the reason for the difficulties to discern any genotype-phenotype correlation in patients carrying mutations in LIPH or the gene encoding the P2RY5 receptor, LPAR6.
In conclusion, our findings extend the spectrum of known LIPH mutations causing hypotrichosis and woolly hair and further support a crucial role of LIPH in hair growth and texture in humans.
Methods
All experiments were performed in accordance with relevant guidelines and regulations and approved by the local ethical board at NIBGE, Faisalabad, Pakistan. Informed consent was obtained from all individuals who participated in this study. We investigated five consanguineous Pakistani families (Family 1–5) segregating autosomal recessive hypotrichosis. None of the families were previously reported. Affected individuals in all five families exhibited typical features of hypotrichosis, but with a variable phenotype. The scalp hair is sparse to absent, eyebrows and eyelashes are normal or short and sparse, and the body hairs are normal or sparse. The main feature of Family 1, 2 and 3 is hypotrichosis without woolly hair, while affected members of Family 4 and 5 exhibit features of woolly hair together with hypotrichosis. Consistent with previous reports, some affected individuals (Family 2 and 5) also show a slight depigmentation of the hair (Figure 1 and Table 1)9,10,11,12. All patients reported normal sweating and no additional ectodermal or physical abnormalities were observed. Blood samples were obtained from affected individuals, siblings and their parents when available. Genomic DNA was extracted from peripheral blood according to standard techniques. We initially analysed all five families for linkage to the three known LAH genes using highly polymorphic microsatellite markers flanking the DSG4 gene locus on chromosome 18q12.1, the LIPH gene locus on chromosome 3q27.2, and the LPAR6 gene locus on chromosome 13q14.11–q21.32, respectively. Two-point logarithm of odds (LOD) scores was calculated using MLINK software from the LINKAGE program package13. All coding and exon-flanking sequences of the LIPH gene were directly sequenced with Big Dye Terminator v3.1 cycle sequencing kit according to manufacturer's protocol (Applied Biosystems, Foster City, CA) and separated on an ABI 3730xl DNA analyzer (Applied Biosystems).
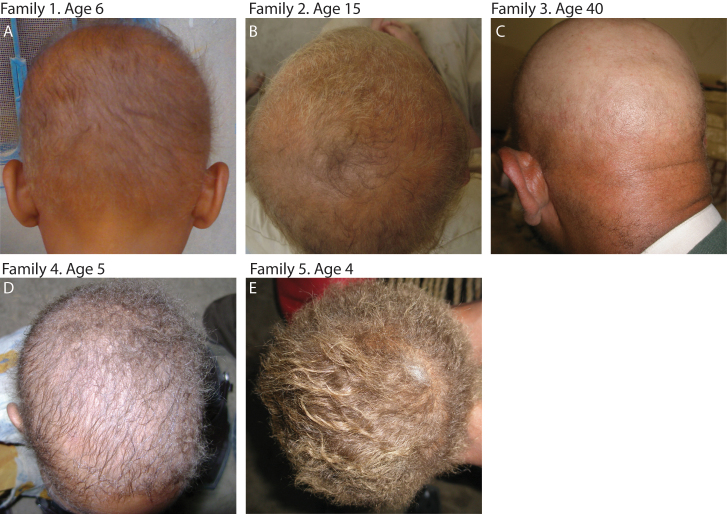
Figure 1. Phenotypic overview of patients from the five families.
(A) Affected male (age 6) member of family 1 showing hypotrichosis (B) Affected male (age 15) member of family 2 showing hypotrichosis and hair depigmentation. (C) Affected male (age 40) member of family 3 showing severe hypotrichosis. (D) Affected female (age 5) member of family 4 showing hypotrichosis and woolly hair. (E) Affected female (age 4) member of family 5 showing mild hypotrichosis, woolly hair and hair depigmentation.
Author Contributions
MT and AA contributed equally to this work. MT, AA and SMB identified, informed, and sampled the patients. ND clinically evaluated the patients. MT, AA and JK performed gene mapping, mutation screening and bioinformatic analysis. JK directed the overall research and designed the study together with ND. JK wrote the manuscript, all author's reviewed the manuscript.
Acknowledgments
This work was supported by grants from the Swedish Research Council (K2010-66X-10829-17-3) and Swedish Links, Uppsala University Hospital, Uppsala University and the Science for Life Laboratory, J.K was supported by the Swedish Society for Medical Research and M.T. and A.A. by the Swedish Institute.
References
- Aslam M. et al. A novel locus for autosomal recessive form of hypotrichosis maps to chromosome 3q26.33-q27.3. J Med Genet 41, 849–52 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafique M. A. et al. A locus for hereditary hypotrichosis localized to human chromosome 18q21.1. Eur J Hum Genet 11, 623–8 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wali A. et al. Localization of a novel autosomal recessive hypotrichosis locus (LAH3) to chromosome 13q14.11-q21.32. Clin Genet 72, 23–9 (2007). [DOI] [PubMed] [Google Scholar]
- Ali G., Chishti M. S., Raza S. I., John P. & Ahmad W. A mutation in the lipase H (LIPH) gene underlie autosomal recessive hypotrichosis. Hum Genet 121, 319–25 (2007). [DOI] [PubMed] [Google Scholar]
- Inoue A. et al. LPA-producing enzyme PA-PLAalpha regulates hair follicle development by modulating EGFR signalling. EMBO J 30, 4248–60 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternack S. M. et al. Novel mutations in the P2RY5 gene in one Turkish and two Indian patients presenting with hypotrichosis and woolly hair. Arch Dermatol Res 301, 621–4 (2009). [DOI] [PubMed] [Google Scholar]
- Jelani M., Wasif N., Ali G., Chishti M. & Ahmad W. A novel deletion mutation in LIPH gene causes autosomal recessive hypotrichosis (LAH2). Clin Genet 74, 184–8 (2008). [DOI] [PubMed] [Google Scholar]
- Shimomura Y., Wajid M., Petukhova L., Shapiro L. & Christiano A. M. Mutations in the lipase H gene underlie autosomal recessive woolly hair/hypotrichosis. Journal of Investigative Dermatology 129, 622–8 (2009). [DOI] [PubMed] [Google Scholar]
- Petukhova L. et al. The effect of inbreeding on the distribution of compound heterozygotes: a lesson from Lipase H mutations in autosomal recessive woolly hair/hypotrichosis. Hum Hered 68, 117–30 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. et al. Mutations in the LPAR6 and LIPH genes underlie autosomal recessive hypotrichosis/woolly hair in 17 consanguineous families from Pakistan. Clin Exp Dermatol 36, 652–4 (2011). [DOI] [PubMed] [Google Scholar]
- Kurban M., Ghosn S., Abbas O., Shimomura Y. & Christiano A. A missense mutation in the P2RY5 gene leading to autosomal recessive woolly hair in a Syrian patient. J Dermatol Sci 57, 132–4 (2010). [DOI] [PubMed] [Google Scholar]
- Horev L., Saad-Edin B., Ingber A. & Zlotogorski A. A novel deletion mutation in P2RY5/LPA(6) gene cause autosomal recessive woolly hair with hypotrichosis. J Eur Acad Dermatol Venereol 24, 858–9 (2010). [DOI] [PubMed] [Google Scholar]
- Lathrop G. M. & Lalouel J. M. Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet 36, 460–5 (1984). [PMC free article] [PubMed] [Google Scholar]