Abstract
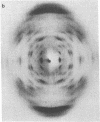
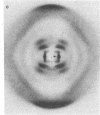
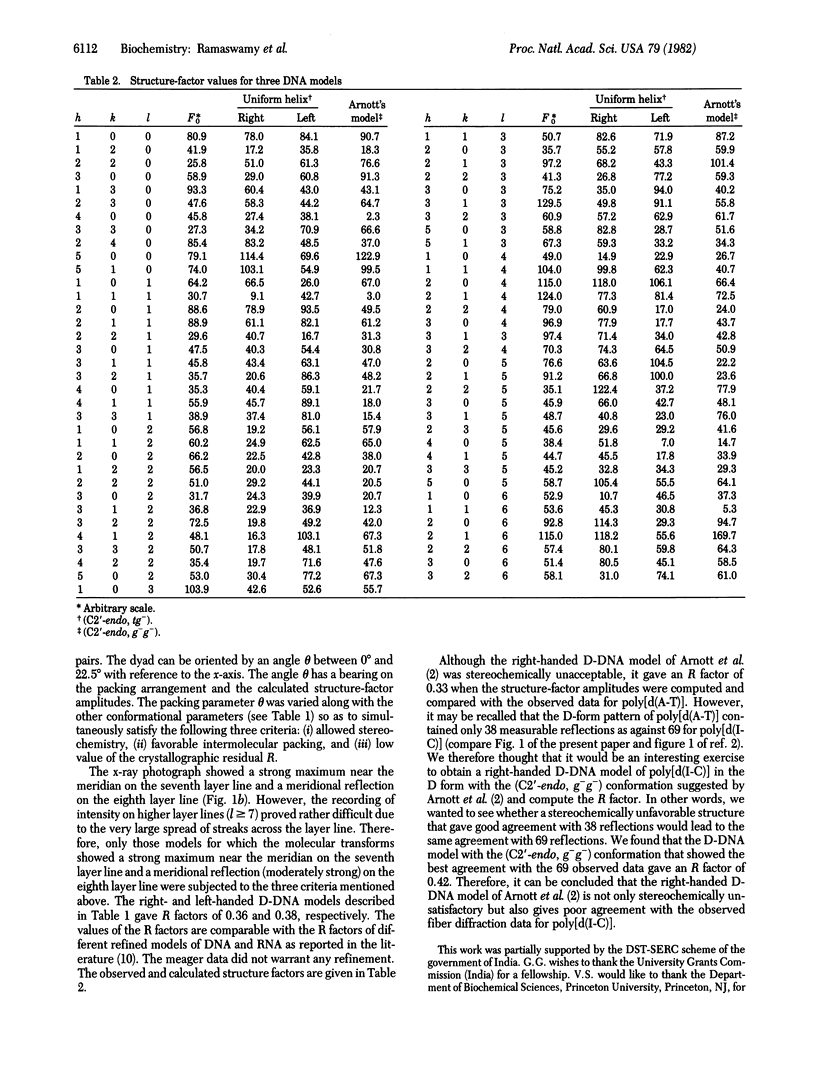
Earlier, we showed that, for the D form (n = 8 and h = 3.03 A, where n is number of nucleotide units per turn and h is height per nucleotide unit) of poly[d(A-T)], both right- and left-handed double helical models are stereochemically satisfactory and give good agreement with the observed fiber diffraction data. It was also noted that the conformations of the right- and left-handed D-DNA models are very similar to those of the right- and left-handed B-DNA models. This observation was consistent with the D leads to B transition in the solid phase. As a continuation of our earlier studies, we have carried out similar experiments with poly[d(I-C)]. We could obtain a crystalline D-form pattern (n = 8, h = 3.13 A) of the fiber at 75% relative humidity (r.h.); the hydrated (r.h. approximately equal to 95%) form of the same fiber gave the classical B-form pattern (n = 10, h = 3.40 A). In the present report, we show that both right- and left-handed double-helical models are consistent with the fiber diffraction data of poly[d(I-C)] in the D-form. Theoretical energy calculations also suggest that the right- and left-handed B- and D-DNA models are almost equally stable. Hence, we conclude that the right- and left-handed double-helical models of poly[d(I-C)] in a given form (B or D) are equally likely and that the fiber diffraction data do not permit discrimination.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Chandrasekaran R., Hukins D. W., Smith P. J., Watts L. Structural details of double-helix observed for DNAs containing alternating purine and pyrimidine sequences. J Mol Biol. 1974 Sep 15;88(2):523–533. doi: 10.1016/0022-2836(74)90499-9. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. Refinement of the structure of B-DNA and implications for the analysis of x-ray diffraction data from fibers of biopolymers. J Mol Biol. 1973 Dec 5;81(2):93–105. doi: 10.1016/0022-2836(73)90182-4. [DOI] [PubMed] [Google Scholar]
- Arnott S. The geometry of nucleic acids. Prog Biophys Mol Biol. 1970;21:265–319. doi: 10.1016/0079-6107(70)90027-1. [DOI] [PubMed] [Google Scholar]
- Gupta G., Bansal M., Sasisekharan V. Conformational flexibility of DNA: polymorphism and handedness. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6486–6490. doi: 10.1073/pnas.77.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui Y., Langridge R., Shortle B. E., Cantor C. R., Grant R. C., Kodama M., Wells R. D. Physical and enzymatic studies on poly d(I-C)-poly d(I-C), an unusual double-helical DNA. Nature. 1970 Dec 19;228(5277):1166–1169. doi: 10.1038/2281166a0. [DOI] [PubMed] [Google Scholar]