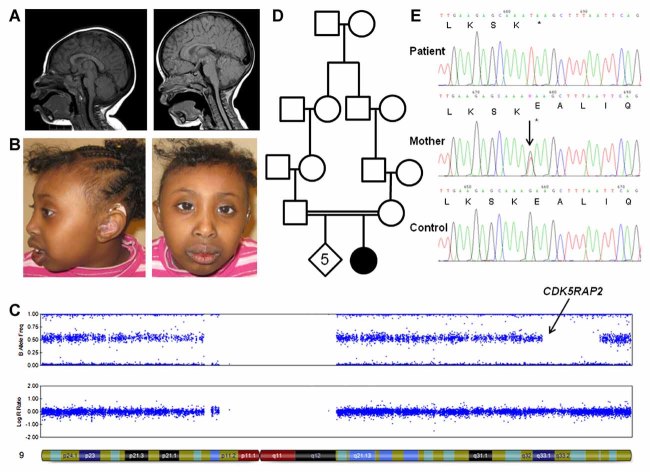
FIG. 1.
A: Sagittal T1SE MRI images of patient at 15 months (on left) compared to a normal age- and gender-matched child (on right), demonstrating cranio-facial disproportion characteristic of microcephaly. B: Photographs taken at age 6 years, shown with parental consent, indicating microcephaly and sloping forehead. C: SNP-array data for chromosome 9 showing a 13.7 Mb region of copy-neutral loss of heterozygosity at chr9:120,050,463–133,809,775 (GRCh19/hg37). In combination with a second region of cnLOH (chr7:9,870,471–27,658,801), the coefficient of inbreeding was estimated to be ∼1/95. D: Simplified pedigree showing that the parents of the patient are second-cousins. Black shading indicates primary microcephaly and hearing loss. The patient is the fifth child in a sibship of six. We note that the male–male link in this consanguineous loop means that the homozygous region on Xq22.3 is unlikely to have come from these great-great-grandparents. E: Sanger sequencing identified a homozygous chr9:123,292,381C>A mutation, inherited from the heterozygous mother. Electropherogram shows sequence on the negative (i.e., coding) strand so mutation appears as G>T and predicts a Glu → STOP codon. DNA from the father and the patient's siblings was not available.