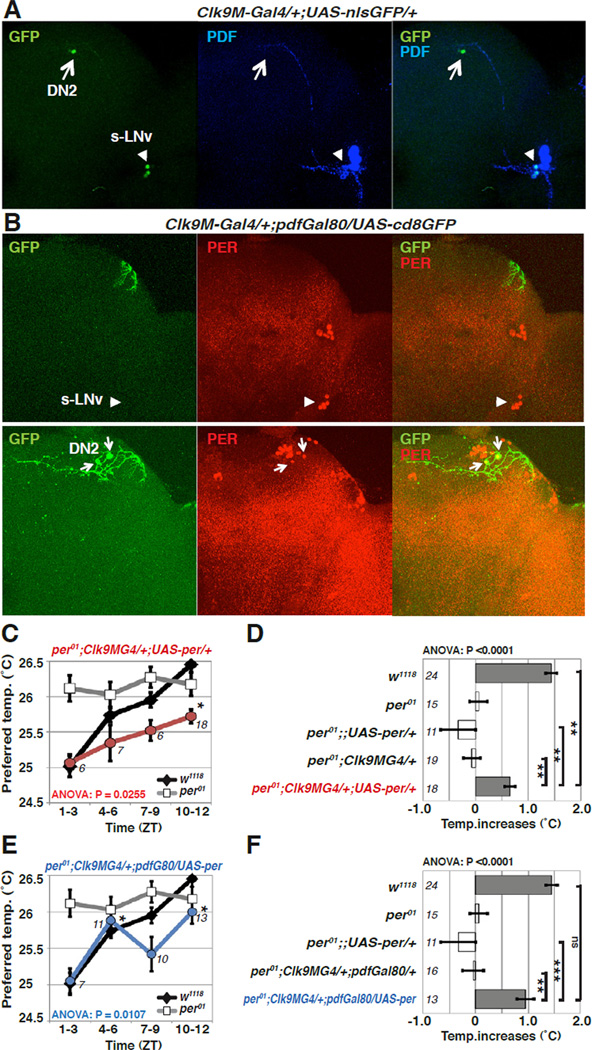
Figure 4. DN2 neurons are sufficient to generate daytime TPR in LD.
(A-B) Clk9M-Gal4; pdf-Gal80 is selectively expressed in DN2s. (A)Clk9M-Gal4::UAS-nlsGFP (nuclear GFP) flies were stained with anti-GFP (green) and anti-PDF (blue). The PDF antibody labels the s-LNv (arrowhead) and l-LNvs neurons. Clk9M-Gal4 labels both DN2 (arrow) and s-LNv neurons (arrowhead). 2 DN2 cells are observed in Clk9MGal4::UAS-nGFP flies, but GFP staining was frequently weaker in one of them.
(B) Clk9M-Gal4, pdf-Gal80/ UAS-cd8GFP flies are stained with anti-GFP and anti-PER. The PER antibody labels circadian pacemaker cells in the brain. pdf-Gal80 eliminates the expression in s-LNv neurons. Clk9M-Gal4, pdf-Gal80/ UAS-cd8GFP is selectively expressed in the DN2 neurons. Upper and lower pictures show different Z-stacks of the same brain.
(C–F) PER expression in DN2 neurons is sufficient to rescue daytime TPR of per01 in LD. The line graphs (C, E) show preferred temperatures of w1118 and rescue fly lines in the null mutant per01 background during the daytime. The same LD daytime data from Figures 1B, and 2F were used for w1118 and per01, respectively. PER expression in Clk9M-Gal4, pdf-Gal80 (E, blue) is sufficient to rescue the per01 mutant phenotype for daytime TPR (ZT1–12), while PER expression in Clk9M-Gal4 (C, red) partially rescues it. The bar graphs (D, F) show the daytime TPR (ZT1–12) for each genotype. ANOVA was performed on the following groups; w1118, per01;;UAS-per/+, per01; Clk9M-Gal4/+, and per01; Clk9M-Gal4/+;UAS-per/+ (D), as well as w1118, per01;;UAS-per/+, per01; Clk9MGal4/+; pdf-Gal80/+, and per01; Clk9M-Gal4/+; pdf-Gal80/UAS-per (F), Tukey-Kramer test comparison to ZT1–3 (C, E) and each control fly line (D, F). ***P<0.001, **P<0.01 or *P<0.05.