Abstract
Morphogenesis of the vertebrate head relies on proper dorsal-ventral (D-V) patterning of neural crest cells (NCC) within the pharyngeal arches. Endothelin-1 (Edn1)-induced signaling through the endothelin-A receptor (Ednra) is crucial for cranial NCC patterning within the mandibular portion of the first pharyngeal arch, from which the lower jaw arises. Deletion of Edn1, Ednra or endothelin-converting enzyme in mice causes perinatal lethality due to severe craniofacial birth defects. These include homeotic transformation of mandibular arch-derived structures into more maxillary-like structures, indicating a loss of NCC identity. All cranial NCCs express Ednra whereas Edn1 expression is limited to the overlying ectoderm, core paraxial mesoderm and pharyngeal pouch endoderm of the mandibular arch as well as more caudal arches. To define the developmental significance of Edn1 from each of these layers, we used Cre/loxP technology to inactivate Edn1 in a tissue-specific manner. We show that deletion of Edn1 in either the mesoderm or endoderm alone does not result in cellular or molecular changes in craniofacial development. However, ectodermal deletion of Edn1 results in craniofacial defects with concomitant changes in the expression of early mandibular arch patterning genes. Importantly, our results also both define for the first time in mice an intermediate mandibular arch domain similar to the one defined in zebrafish and show that this region is most sensitive to loss of Edn1. Together, our results illustrate an integral role for ectoderm-derived Edn1 in early arch morphogenesis, particularly in the intermediate domain.
Keywords: neural crest cell, conditional knockout, mouse, endothelin, craniofacial
INTRODUCTION
Cranial neural crest cells (NCCs), a multipotent cell population generated at the interface between the non-neural ectoderm and neural tube, form most of the craniofacial structures, including bone, cartilage, connective tissue and portions of the cranial nerves (Bronner-Fraser, 1995; Le Douarin et al., 1993). During development of the upper and lower jaws, NCCs migrate ventrally around the embryo and populate the pharyngeal arches, initiating a mesenchymal differentiation program (Le Douarin, 1982; Lumsden et al., 1991). Dorsal-ventral (D-V) patterning of this preskeletal NCC-derived mesenchyme is dependent on signals generally arising from the surrounding cell types within the arches (Chai and Maxson, 2006; Clouthier et al., 2010). Many of these signals act in both instructive and inhibitory manners that enforce sub-domains within the pharyngeal arches necessary for the development of regionally restricted bone, cartilage and connective tissue structures.
One of the key initiators of D-V patterning within the first mandibular arch NCCs is endothelin-1 (Edn1) (Clouthier et al., 2010), which is expressed by cells in the pharyngeal arch environment, including the ventral arch ectoderm, core paraxial mesoderm and pharyngeal arch endoderm (Clouthier et al., 1998; Maemura et al., 1996; Yanagisawa et al., 1998a; Yanagisawa et al., 1998b), while its cognate receptor, the endothelin-A receptor (Ednra) is expressed by NCCs that populate the pharyngeal arches (Clouthier et al., 1998; Yanagisawa et al., 1998a). Disruption of Ednra signaling leads to loss of NCC polarity and thus an expansion of the dorsal (proximal in mouse) identity into the ventral (distal in mouse) arch. This includes homeotic transformation of lower jaw structures and middle ear structures into more maxillary-like structures (Kimmel et al., 2003; Nair et al., 2007; Ozeki et al., 2004; Ruest et al., 2004; Sato et al., 2008) and inappropriate ventral expression of jag1b and hey1 (the latter induced by Jagged-Notch signaling (Zuniga et al., 2010)).
Since Edn1 is expressed in multiple domains in the developing arches, a fundamental question remains concerning the contribution of each domain to patterning. Studies in zebrafish have supported a role for Edn1 expressed in the ectoderm, but the relevance of these findings to the mammalian system has been confounded by the presence of two copies of Ednra in the zebrafish as well as by differences in the morphogenesis of the lower jaw between fish and mammals. In particular, Edn1 is most important for the intermediate domain in the fish, which will form the joint between Meckel’s cartilage and the palatoquadrate, whereas this latter structure is not present in mammals. Although an intermediate domain has not previously been noted in mammals, pharmacological antagonism of Ednra signaling resulted in disproportionate changes in gene expression within the central mandibular arch (Clouthier et al., 2003). In this study, we have taken advantage of Cre/loxP technology to disrupt Edn1 expression in the first arch ectoderm, endoderm or mesoderm. We find that changes in facial patterning only occur when Edn1 is inactivated in the ectoderm. In addition, this approach has unexpectedly allowed us to define the intermediate domain of the mandibular arch in mice and identify its structural significance. Our findings point to this domain as a crucial signaling center in mammalian lower jaw development.
MATERIALS AND METHODS
Mice
The generation, characterization and genotyping protocols for Edn1flox/flox (Edn1fl/fl) (Kisanuki et al., 2010), Foxg1-Cre (Hebert & McConnell, 2000), Myf5-Cre (Tallquist et al., 2000; Jackson Laboratories strain B6.129S4-Myf5tm3(cre)Sor/J), Foxa2mcm (Park et al., 2008) and R26R (Soriano, 1999) mice have been previously described. The Crect transgene is comprised of an ectodermal enhancer of Tfap2a driving Cre expression in the ectoderm (Forni et al., 2011).
Generation of mutant embryos
To generate conditional knockout embryos, Edn1fl/+;Cre mice were bred with Edn1fl/fl mice to generate Edn1fl/fl;Foxg1-Cre, Edn1fl/fl;Myf5-Cre, Edn1fl/fl;Foxa2mcm and Edn1fl/fl;Crect embryos. Because the Foxa2-Cre construct is tamoxifen-inducible, embryonic day (E) 6.5 pregnant female mice received tamoxifen. To accomplish this, tamoxifen was first dissolved in 100% ethanol at 10 mg/100 µl. Mineral oil was added to obtain a final concentration of 10 mg/ml. After sonicating for 45 min, progesterone was added at 5 mg/ml (which can limit fetal toxicity of tamoxifen (Jackson Laboratories)) and then the tamoxifen/progesterone mixture used immediately. Pregnant mice were weighed and then injected intraperitoneally with 75 µg per gram body weight of the tamoxifen/progesterone mixture.
Confirmation of allele recombination
Cre-mediated recombination analysis of the Edn1flox allele was performed as described (Kisanuki et al., 2001) using DNA extracted with the Puregene Tissue Core Kit A (Gentra) from E9.0 wild type and mutant embryo pharyngeal arches. Briefly, extracted DNA was measured using a Nanodrop ND-1000 spectrophotometer (Nanodrop) and 20 ng/µl of genomic DNA was added to each PCR reaction. The reactions, performed with the same settings previously described (Kisanuki et al., 2001), generate a 300 bp product for the recombined allele.
β-galactosidase staining
Whole-mount staining was performed as previously described (Ruest et al., 2003). Briefly, embryos were collected between embryonic day (E) 8.0 to E10.5, fixed in 4% paraformaldehyde (PFA) on ice for 1 hour then processed for β-galactosidase staining overnight at room temperature. After staining, embryos were rinsed and photographed on an Olympus SZX12 stereomicroscope fitted with a DP11 digital camera. For more detailed analysis of staining, these stained E9.0 embryos were embedded in Paraplast Plus tissue embedding medium and sectioned at 12 µm. Sections were collected on Pluscoated slides (Fisher) and counterstained with nuclear fast red as previously described (Ruest et al., 2003). Sections were analyzed and photographed using an Olympus BX51 compound microscope fitted with a DP71 digital camera.
Skeleton staining
Cartilage staining of E13.5 and E14.5 embryos with alcian blue was performed as previously described (Clouthier et al., 1998). Skeletal staining of E18.5 embryos with alizarin red (bone) and alcian blue (cartilage) was performed as previously described (Ruest et al., 2004). Stained embryos were analyzed and photographed using the Olympus SZX12 stereomicroscope as described above.
Whole-mount in situ hybridization
Whole-mount single probe in situ hybridization (ISH) analysis was performed as described previously (Clouthier et al., 1998) using digoxigenin (DIG)-labeled antisense cRNA riboprobes against Dlx2, Dlx3, Dlx5, Hand2 and Goosecoid. For dual probe whole-mount in situ hybridization, fluorescein-labeled antisense cRNA riboprobes against Dlx3 and Dlx5 were used in combination with DIG-labeled riboprobes against Bapx1/Nkx3.2. Processing and hybridization was identical to that of the single probe protocol, though maleic acid buffer plus Tween-20 (MABT) was used for embryo washes. Embryos were blocked with MABT + 2% blocking reagent (Roche) + 20% sheep serum, incubated overnight with 1:2,000 anti-fluorescein-AP antibody (Roche) in blocking solution, washed with MABT and developed with magenta-phos (Biosynth) in pH 8.5 NTMT. The first AP enzyme was killed by incubating developed embryos in 65°C MABT for 1 hour. After this step, embryos were blocked again, incubated with 1:2,000 anti-digoxigenin-AP antibody (Roche) in blocking solution and developed with BCIP (Roche) in pH 9.5 NTMT. Embryos were photographed using an Olympus SZX12 microscope as described above.
RESULTS
Early expression domains of Foxg1-Cre, Myf5-Cre, Foxa2mcm and Crect
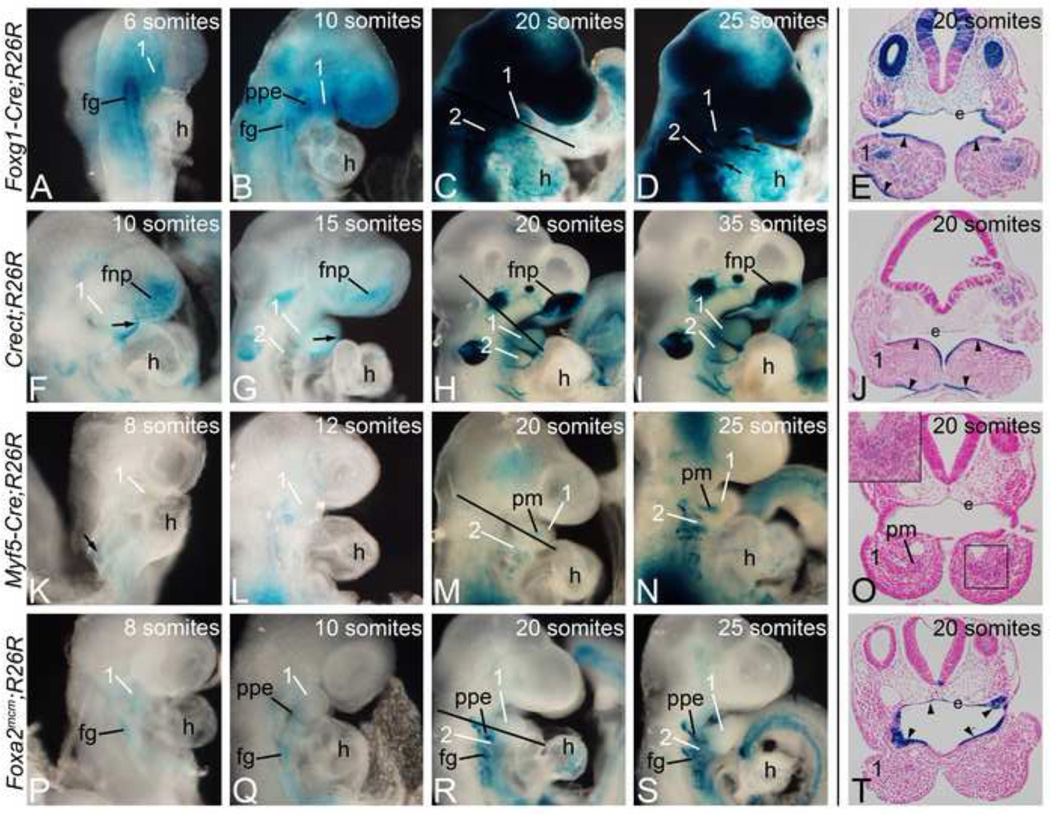
To inactivate Edn1 in a tissue-specific manner, we used four Cre transgenic mouse strains: Foxg1-Cre (endoderm/mesoderm/ectoderm) (Hebert & McConnell, 2000), Crect (ectoderm) (Forni et al., 2011), Myf5-Cre (mesoderm)(Tallquist et al., 2000) and Foxa2mcm (endoderm) (Park et al., 2008). To first confirm that Cre-mediated loxP recombination occurred in a spatio-temporal manner sufficient to delete Edn1 prior to or during Edn1 function (which is between E8.5 and E9.0 (Ruest and Clouthier, 2009)). Cre animals were crossed into the R26R strain (Soriano, 1999). Embryos were harvested between E8.0 (6 somites) and E10.5 (35 somites) and stained for beta-galactosidase (β-gal) activity. In Foxg1-Cre embryos, Cre sequence has been inserted into the Foxg1 locus (Hebert & McConnell, 2000). In R26R;Foxg1-Cre embryos, β-gal activity was observed by 6 somites (E8.0) in the endoderm/foregut region and the first arch (Fig. 1A). At 10 and 20 somites (E8.5 and E9.0, respectively), foregut staining was still present, with staining also present in the pharyngeal arches and frontonasal prominence (Fig. 1B, C). Transverse sections through these stained embryos illustrated that β-gal staining was confined to the arch ectoderm (arrowheads), pharyngeal pouch endoderm, and core paraxial mesodermal cells (Fig. 1E). Staining at 25 somites (E9.5; Fig. 1D) appeared similar to that observed at 20 somites. Based on this staining, the Foxg1-Cre strain would likely result in an almost complete inactivation of Edn1 within the arches, providing a good control when examining the phenotypes resulting from the use of our other more tissue-specific Cre strains.
Figure 1. Foxg1-Cre, Crect, Myf5-Cre, and Foxa2mcm activity.
Cre animals were crossed into the R26R Cre reporter strain. Embryos were harvested between E8.0 (6 somites) and E10.5 (35 somites) and stained for β-gal activity. A–D. Cre activity in R26R;Foxg1-Cre embryos was found in endodermal (foregut (fg) and pharyngeal pouch endoderm (ppe)) and ectodermal structures. E. Transverse section through the embryo in C (20 somites) illustrates staining in the arch ectoderm (arrowheads) and endoderm (e), with staining also observed in the paraxial core mesoderm. F–I. β-gal activity in R26R;Crect embryos was found in the ectoderm of the arches (arrow) and in the frontonasal prominence (fnp). J. Transverse section of embryo in H showing arch ectodermal staining at 20 somites (arrowheads). K–N. R26R;Myf5-Cre embryos showed mesodermal staining that at 20 somites could be seen in the paraxial core mesoderm (pm). O. A transverse section through the arches in M illustrated that the staining is restricted to the core mesoderm. (inset). P–S. β-gal activity in Foxa2mcm;R26R embryos starts at E8.0 following tamoxifen injection at E6.5, with endodermal structures labeled (ppe, fg). T. Sections through the first pharyngeal pouch endoderm (e) at 20 somites shows specific β-gal activity (arrowheads). 1, mandibular pharyngeal arch; 2, second pharyngeal arch; h, heart.
We next analyzed β-gal staining in R26R;Crect embryos. This strain expresses Cre under the regulation of a Tfap2 α ectodermal-specific enhancer (T. Williams, manuscript in preparation). Cre activity was detected in the first pharyngeal arch ectoderm (arrow in Fig. 1F, G) and frontonasal prominence by 10 somites (E8.5; Fig. 1F), with the staining intensity increasing by 20 somites (E9.0; Fig. 1H). β-gal staining was also present in the otic vesicle, trigeminal ganglion and optic placode between 15 and 20 somites (Fig. 1G–I). β-gal staining extended to the ectoderm of more caudal pharyngeal arches by 20 somites (E9.0; Fig. 1H). Transverse sections of these embryos showed that most arch ectodermal cells were β-gal-positive (Fig. 1J). This pattern remained unchanged at 35 somites (E10.5) (Fig. 1I).
In the Myf5-Cre strain, a Cre cDNA has been inserted into the Myf5 locus so that Cre expression is under control of Myf5 regulatory sequences (Tallquist et al., 2000). β-gal staining in R26R;Myf5-Cre embryos was observed starting at 8 somites (E8.0) in the somatic mesoderm (Fig. 1K). At 12 somites (E8.5), we detected β-gal staining adjacent to the pharyngeal arches (Fig. 1L). By 20 somites (E9.0), this staining was restricted to the core mesoderm in the pharyngeal arches (Fig. 1M). Transverse sections through the arch of this embryo illustrated this restriction to the core mesoderm, though staining was faint (Fig. 1O and inset in Fig. 1O). By 25 somites (E9.5), the core mesoderm staining in the pharyngeal arches was robust (Fig. 1N).
Foxa2mcm animals express a tamoxifen-inducible Cre cassette from the Forkhead box A2 (Foxa2) locus in the early embryos endoderm (Park et al., 2008). Following injection of pregnant R26R;Foxa2mcm females with tamoxifen at a stage equivalent to E6.5, embryos were collected between 8 and 25 somites (E8.0 to E9.5). At 8 somites (E8.0), β-gal staining was observed in the foregut endoderm (Fig. 1P). By 10 somites, this staining was more intense (Fig. 1Q–S). β-gal staining was also detected in the first pharyngeal pouch endoderm (Fig. 1Q). By 20 somites (E9.0), this endodermal pouch staining was very intense (Fig. 1R). Transverse sections through the pharyngeal arch illustrated that expression was confined to the arch endoderm (Fig. 1T). Expression was unchanged at 25 somites (E9.5; Fig. 1R, S).
Cre expression drives recombination of the conditional Edn1 locus
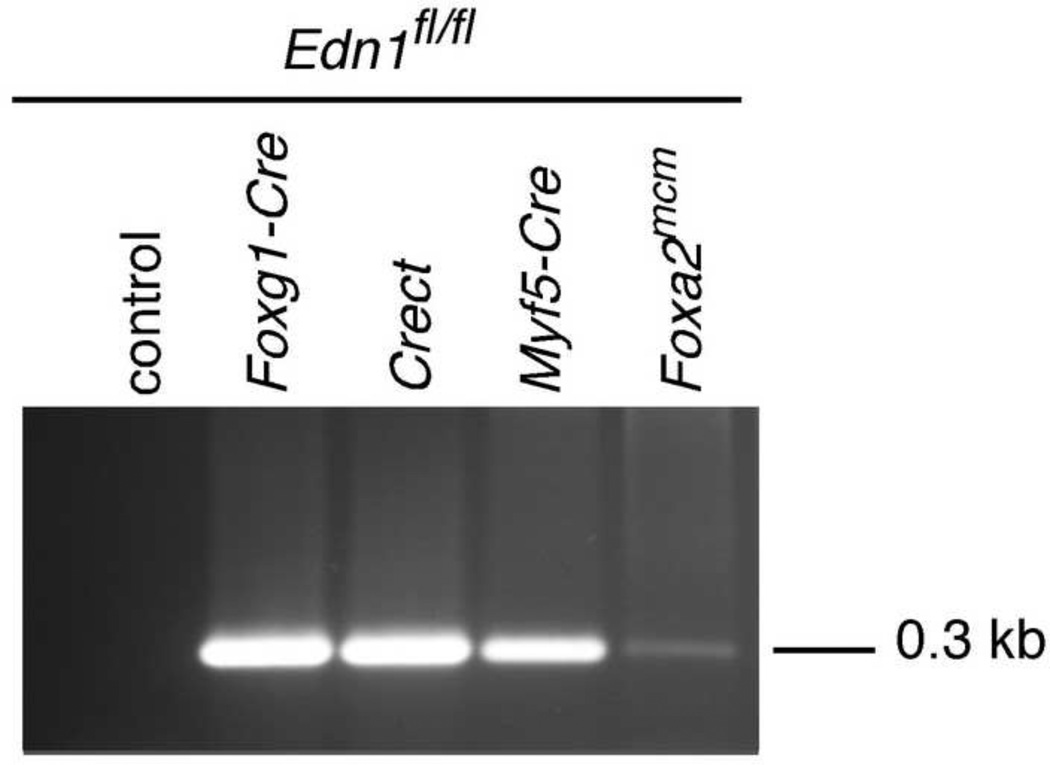
To first confirm that our Cre strains could delete our conditional Edn1 allele, we crossed mice containing a conditional allele of the Edn1 gene (Edn1fl/fl) with the different Cre strains described above. Mandibular arches were collected from E9.0 embryos, with genomic DNA extracted and used in a recombination-specific PCR using primers that only detected the recombined Edn1 allele (Kisanuki et al., 2001). While Ednra signaling is required as early as E8.5, maximum sensitivity to loss of Ednra signaling occurs at E9.0 (Ruest and Clouthier, 2009). In our recombination assay, primers flanking the deleted region of the allele were used for PCR; recombination of the floxed allele resulted in the presence of a 300 bp band. This band was observed in all Edn1fl/fl;Cre embryos, indicating that the Edn1 conditional allele was recombined by all four Cre strains (Fig. 2). This recombination band was robust for all samples except that from the mandibular arch of Edn1fl/fl;Foxa2mcm embryos. Since robust recombination was observed in R26R;Foxa2mcm embryos (Fig. 1R, T) and each reaction contained an identical amount of input DNA, the low intensity of the recombined band in the Edn1fl/fl;Foxa2mcm sample likely reflects the amount of endoderm that existed in the excised mandibular arch.
Figure 2. Analysis of Cre-mediated recombination of the Edn1 gene in Edn1fl/fl;Cre mice.
Genomic DNA was extracted from E9.0 mouse embryo mandibular arches. Lanes 2–5 show the 300-bp band for the recombined allele. Wild-type animals show no band (lane 1).
Ectodermal loss of Edn1 leads to changes in craniofacial bone and cartilage development
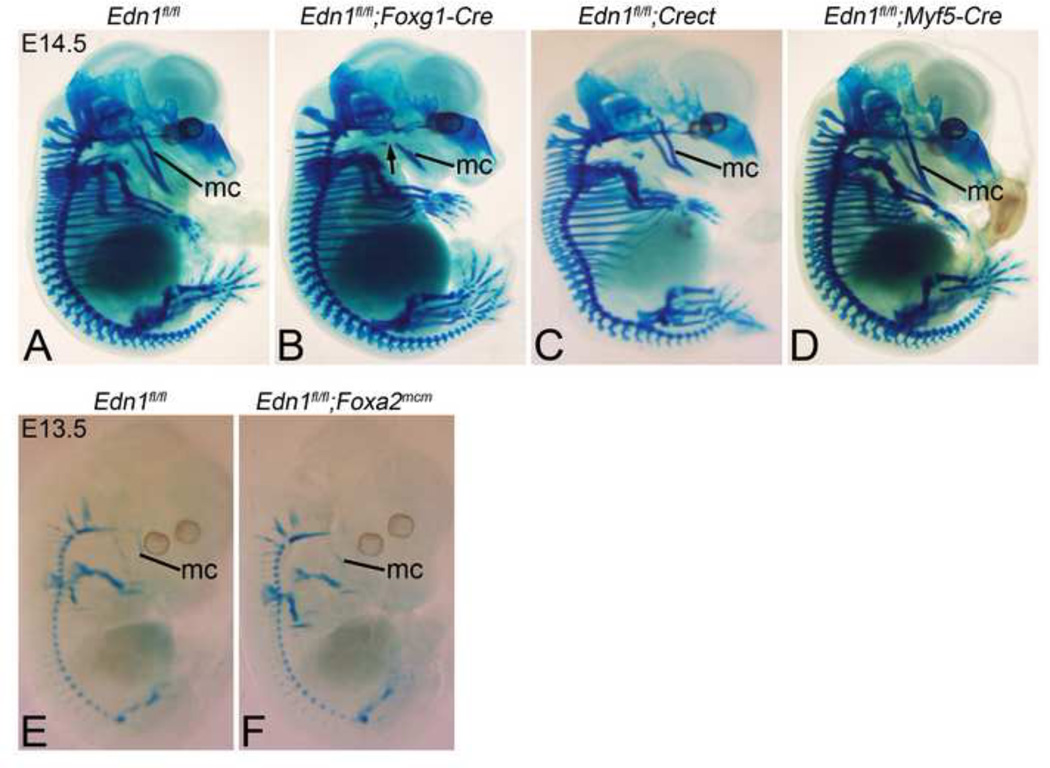
To determine how tissue-specific loss of Edn1 affected D-V patterning, we began our analysis of Edn1 conditional mutants by examining E13.5 or 14.5 conditional knockout embryos for changes in cartilage development, since this is one of the earliest structural changes observed in endothelin-family knockouts in mice and fish (Clouthier et al., 1998; Miller et al., 2000). Deletion of Edn1 in the pharyngeal arch ectoderm, pharyngeal pouch endoderm and core paraxial mesoderm of E14.5 Edn1fl/fl;Foxg1-Cre embryos resulted in hypoplasia of Meckel’s cartilage (Fig. 3B). There was also a gap in Meckel’s cartilage at its proximal end where it failed to articulate with the malleus (Fig. 3B). In addition, the hyoid bone appeared more rostral in the pharynx of Edn1fl/fl;Foxg1-Cre embryos (Fig. 3B), a finding observed in Ednra, Edn1 and Ece1 mutant embryos (Clouthier et al., 1998; Kurihara et al., 1994; Yanagisawa et al., 1998b). However, the overall phenotype was less severe than observed in Edn1−/− mutants (Kurihara et al., 1994). When Edn1 was only deleted in the pharyngeal arch ectoderm of E14.5 Edn1fl/fl;Crect embryos, Meckel’s cartilage appeared slightly shortened, though its articulation with the malleus was normal (Fig. 3C). The hyoid bone positioning again appeared more rostral, but not to the same extent as observed in Edn1fl/fl;Foxg1-Cre embryos (Fig. 3B). In contrast, Meckel’s cartilage was unaffected in E14.5 Edn1fl/fl;Myf5-Cre embryos (Fig. 3D). While we also attempted to examine changes in cartilage structures in E14.5 Edn1fl/fl;Foxa2mcm embryos, administration of tamoxifen at E6.5 (shown to achieve optimal gene recombination at E8.5 (Park et al., 2008) resulted in embryo lethality by E14.5, a general problem with tamoxifen administration before E8.5 (Hayashi and McMahon, 2002). However, we were able to collect E13.5 Edn1fl/fl;Foxa2mcm embryos; in these embryos, the development of Meckel’s cartilage appeared normal in comparison to control embryos (Fig. 3E, F).
Figure 3. Analysis of early skeletal development in Edn1 conditional mutant embryos.
E14.5 (A–D) and E13.5 (E,F) control and mutant embryos stained with alcian blue to visualize cartilage. A–D. Lateral view displaying hypoplastic Meckel’s cartilage (mc) in Edn1fl/fl;Foxg1-Cre (B) and Edn1fl/fl;Crect (C) embryos. Note also the interruption of Meckel’s cartilage in the Edn1fl/fl;Foxg1- Cre embryo (arrow in B) and the altered morphology in the Edn1fl/fl;Crect embryo (arrow in C). Edn1fl/fl;Myf5-Cre embryos (D) appear normal. E–F. Lateral view of Edn1fl/fl (E) and Edn1fl/fl;Foxa2mcm (F) embryos at E13.5, showing no apparent change in Meckel’s cartilage morphology in the mutant.
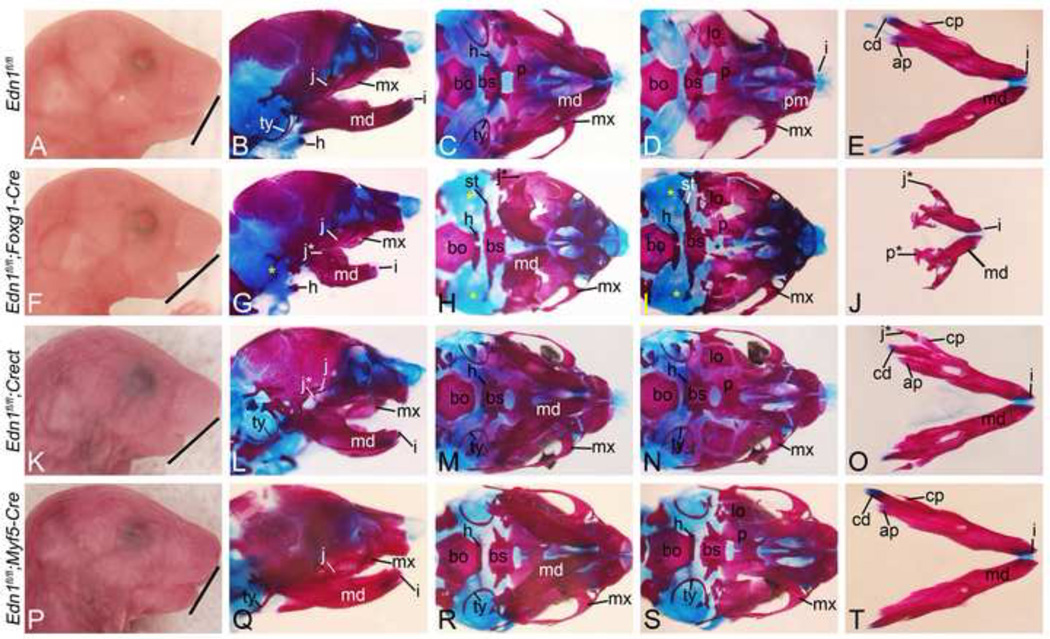
To further examine how tissue-specific Edn1 loss affected skull development, we examined bone development in conditional knockout embryos. In E18.5 Edn1fl/fl;Foxg1-Cre embryos, the lower jaw was shortened and contained mystacial vibrissae on the ectoderm (Fig. 4F and Supp. Fig. 1E). The presence of mystacial vibrissae on the ectoderm is observed in Ednra, Edn1, Dlx5/Dlx6 and Hand2 mutants (Barron et al., 2011; Depew et al., 2002; Ozeki et al., 2004; Ruest et al., 2004). Skeleton preparations of these embryos showed that the mandible was hypoplastic and malformed (Fig. 4G, J), though still retained a mandibular shape, suggesting that patterning of the distal arch mesenchyme was at least partially correct. This is contrast to Edn1−/− embryos, in which the mandible undergoes a homeotic transformation into a maxilla-like structure (Ozeki et al., 2004). However, duplicated palatine (p*) and jugal (j*) bones were present in the lower jaw (Fig. 4G, J), suggesting that D-V patterning of NCC-derived mesenchyme was at least partially disrupted. In addition, tympanic ring bones were absent (yellow asterisks in Fig. 4G–I), the masseter muscle was dysmorphic (Supp. Fig. 1F) and an ectopic bone extended from the gonial bones to the pterygoid bones (Fig. 4I). This ectopic bone, while not present in Edn1−/− embryos (Ozeki et al., 2004), is observed in both Ednrafl/fl;Wnt1-Cre embryos (Abe et al., 2007) and Ednra−/− < − > +/+ chimeric embryos (Clouthier et al., 2003) as well as in embryos from mothers treated for varying times with an Ednra antagonist (Ruest and Clouthier, 2009).
Figure 4. Characterization of cranial features in Edn1fl/fl conditional knockout embryos at E18.5.
Gross morphology and alizarin red (bone) and alcian blue (cartilage)-staining of Edn1fl/fl (AE), Edn1fl/fl;Foxg1-Cre (F–J), Edn1fl/fl;Crect (K–O) and Edn1fl/fl;Myf5-Cre (P–T) embryos. B, G, L, Q. Lateral view. C, H, M, R. Ventral skull view with mandible. D, I, N, S. Ventral view with mandible removed. E, J, O, T. Ventral aboral view of dissected mandibles. A–E. Normal craniofacial structures of an E18.5 control mouse embryo. F–J. Edn1fl/fl;Foxg1-Cre embryos have a recessed lower jaw (note angle of line between A and F), deformed mandible (md) (G, H and J), absent tympanic rings (ty) (yellow * in G–I), an abnormal bone strut (st) extending from the middle ear region towards the basisphenoid (bs) (H–I), a palatal cleft (black * in I) and duplication of jugal (j*) (G, H and J) and palatine (p*) bones (J). K–O. Edn1fl/fl;Crect embryos have a recessed mandible (note angle of line between A and K) and duplicated jugal bones (L and O). P–T. Edn1fl/fl;Myf5-Cre embryo craniofacial structures are normal. ap, angular process; bo, basisoccipital; cd, condyloid process; cp, coronoid process; h, hyoid; i, incisors; j, jugal; lo, lamina obturans; mx, maxilla; p, palatine.
In E18.5 Edn1fl/fl;Crect embryos, the retrognathia observed (Fig. 4K) was similar to that observed in Edn1fl/fl;Foxg1-Cre embryos (Fig. 4F), and mystacial vibrissae were also present in both conditional knockout embryos (Supp. Fig. 1H). However, bone and cartilage staining of Edn1fl/fl;Crect embryos revealed that the mandible bone was less dysmorphoic (Fig. 4L, M, O) than that of Edn1fl/fl;Foxg1-Cre embryos (Fig. 4G, H, I). Appearing only slightly shorter, the primary finding was the presence of duplicated jugal bones (j* - Fig. 4L, O). All other mandibular first arch-derived bones were normal (Fig. 4L–O), though a cleft secondary palate was observed in some mutant embryos (Supp. Fig. 1H, I). In addition, the aberrant bone observed Edn1fl/fl;Foxg1-Cre embryos (Fig. 4H, I) was not observed in Edn1fl/fl;Crect embryos (Fig. 4N). In contrast, E18.5 Edn1fl/fl;Myf5-Cre (Fig. 4P–T) embryos did not have any gross or skeletal changes in the lower jaw. In addition, these homozygous mutant mice were viable and fertile (data not shown). As described above, embryonic lethality at E14.5 of all embryos following tamoxifen administration prevented skeletal analysis of Edn1fl/fl;Foxa2mcm embryos.
Disruption of Edn1 in the ectoderm causes gene expression changes
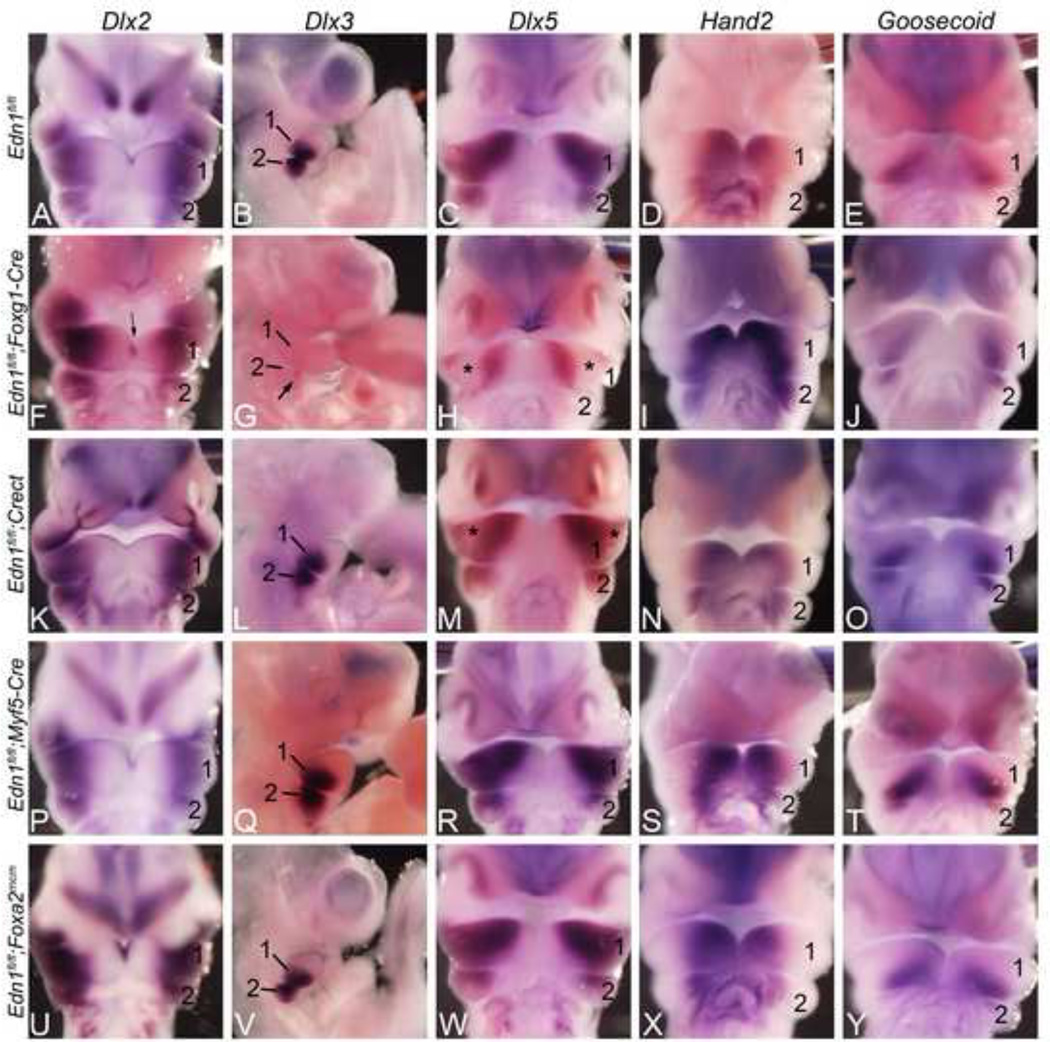
To determine the molecular basis of these observed skeletal changes, we examined the expression of specific genes previously identified as being either induced or repressed downstream of Ednra signaling and crucial for D-V patterning of the mandibular arch. In E10.5 control embryos, Dlx2 expression was observed in the proximal mandibular arch mesenchyme and along the ectoderm overlying the distal mandibular arch (Fig 5A). In E10.5 Edn1fl/fl;Foxg1-Cre embryos, Dlx2 expression was disrupted along most of the distal ectoderm of the mandibular arch (Fig. 5F) similar to that seen in Ednra mutants (Ruest et al., 2004). Unlike in Ednra mutant embryos, mesenchymal expression was not upregulated distally (Fig. 5F). Dlx3 expression was completely lost in the mandibular arch mesenchyme and overlying ectoderm and reduced in the second arch (Fig. 5G and data not shown), again matching the pattern observed in Ednra mutant embryos (Ruest et al., 2004). Dlx5 expression was also disrupted, but only in a band spanning the central mandibular arch (compare Fig. 5C, 5H). Hand2 expression appeared unchanged (Fig. 5I), as did the expression of Goosecoid (Fig. 5J), a gene whose arch expression requires Hand2 (Barron et al., 2011); (Kimmel et al., 2003). Matching the milder phenotype, only Dlx5 expression was disrupted in E10.5 Edn1fl/fl;Crect embryos (Fig. 5M). However, as observed for Edn1fl/fl;Foxg1-Cre embryos, this disruption was only observed in a band spanning the central mandibular arch. Expression of these markers was not disrupted in E10.5 in Edn1fl/fl;Myf5-Cre (Fig. 5P–T) or Edn1fl/fl;Foxa2mcm (Fig. 5U–Y) embryos.
Figure 5. Whole-mount in situ hybridization (ISH) analysis in E10.5 Edn1fl/fl conditional knockout embryos.
Ventral view of embryos after ISH for Dlx2 (A, F, K, P, U), Dlx5 (C, H, M, R, W), Hand2 (D, I, N, S, X) and Goosecoid (E, J, O, T, Y). The heart has been removed to aid in visualization. B, G, L, Q, V show a lateral view of embryos after ISH for Dlx3. A–E. Expression in E10.5 control embryos. F–J. In Edn1fl/fl;Foxg1-Cre embryos, Dlx2 expression was disrupted in the distal ectoderm of the mandibular first pharyngeal arch (arrows in F). Dlx3 expression was almost completely lost in the first pharyngeal arch and reduced in the second pharyngeal arch (arrow in G). Dlx5 expression was disrupted, but only in a band across the central mandibular arch (asterisks in H). Hand2 (I) and Goosecoid (J) expression appeared unaffected. K–O. Dlx5 expression in Edn1fl/fl;Crect embryos was also disrupted (asterisks in M).Expression of other genes examined was unaffected. P–Y. The expression of all five genes was unaffected in Edn1fl/fl;Myf5-Cre (P–T) and Edn1fl/fl;Foxa2mcm (U–Y) embryos. 1, mandibular pharyngeal arch; 2, second pharyngeal arch.
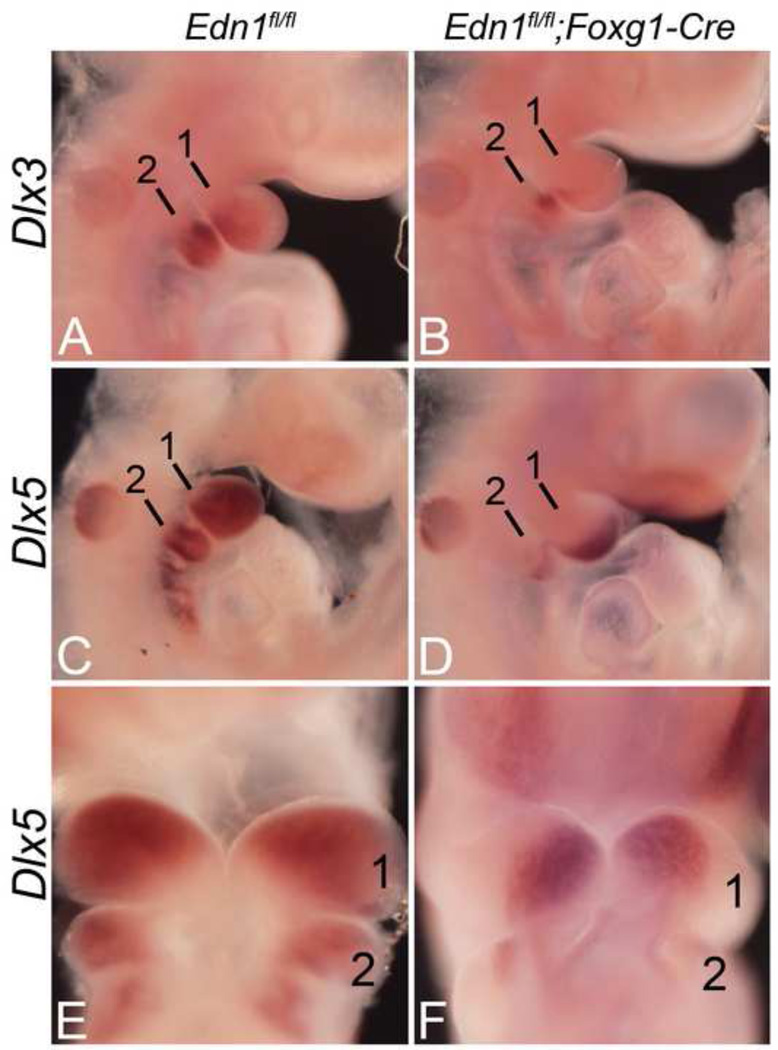
While the changes in the expression of both Dlx3 and Dlx5 were profound in Edn1fl/fl;Foxg1-Cre embryos, these changes could result from either failure to initiate expression or failure to maintain expression. To determine which of these was occurring, we examined the expression of both genes in E9.5 Edn1fl/fl;Foxg1-Cre embryos. When compared to control embryos (Fig 6A), Dlx3 expression in mutants was almost completely absent, suggesting that its expression was never induced (Fig. 6B). While Dlx5 expression was present, expression was confined to the distal half of the mandibular arch (Fig. 6D, F). Expression was absent in the proximal half of the arch, suggesting that the observed expression of Dlx5 in this region in E10.5 Edn1fl/fl;Foxg1-Cre embryos is induced by a different signaling pathway.
Figure 6. Whole-mount in situ hybridization (ISH) analysis in E9.5 Edn1fl/fl;Foxg1-Cre embryos.
Lateral (A–D) and ventral (E–F) views of embryos after ISH for Dlx3 and Dlx5. A–B. Dlx3 expression was almost completely absent in the first arch of E9.5 Edn1fl/fl;Foxg1-Cre embryos (B). C–F. Although Dlx5 expression was present in Edn1fl/fl;Foxg1-Cre embryos, expression was confined to the distal half of the mandibular arch (D, F). 1, mandibular pharyngeal arch; 2, second pharyngeal arch
Disruption of Dlx5 gene expression demarcates the intermediate domain in the mouse mandibular arch
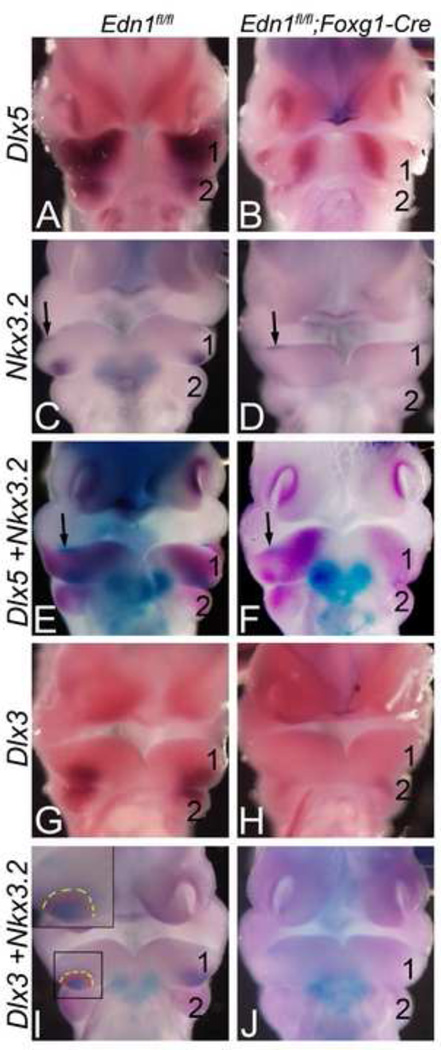
Our in situ hybridization results illustrate that Dlx5 expression in both E10.5 Edn1fl/fl;Foxg1-Cre and Edn1fl/fl;Crect embryos is only affected in a narrow band in the central arch (Fig 5H, M, Fig. 7B, F and Fig. 8B, F). Since this area corresponds spatially to the intermediate domain of the zebrafish mandibular arch, we performed additional analysis to define the gene expression pattern within this region, first examining gene expression changes in Edn1fl/fl;Foxg1-Cre embryos. Nkx3.2 (Bapx1) is the primary molecular marker of the intermediate domain in zebrafish (Miller et al., 2003; Talbot et al., 2010) and a key molecule required for zebrafish jaw joint development (Miller et al., 2003). In E10.5 control embryos, Nkx3.2 expression was confined to a caudal region in the central mandibular arch mesenchyme in addition to a small region along the rostral arch ectoderm (Fig. 7C). In E10.5 Edn1fl/fl;Foxg1-Cre embryos, expression in the caudal mesenchyme was downregulated, though arch ectoderm expression was unaffected. To confirm that the Nkx3.2 expression domain correlated with the domain in which Dlx5 expression was lost, we performed double in situ hybridization analysis. In E10.5 control embryos, Nkx3.2 mesenchyme expression was contained within the Dlx5 expression domain (Fig. 7E). In E10.5 Edn1fl/fl;Foxg1-Cre embryos, the remaining ectodermal Nkx3.2 expression domain was centered in the domain in which Dlx5 was absent (Fig. 7F), strongly suggesting that this Dlx5-free domain is the Nkx3.2 positive intermediate domain.
Figure 7. The Nkx3.2 and Dlx5 expression domains overlap in the mandibular arch and demarcate the intermediate domain.
Ventral view of E10.5 Edn1fl/fl;Foxg1-Cre embryos after single (A–D; G–H) or double (E–F; I–J) ISH for Dlx3, Dlx5 and Nkx3.2. A–B. Dlx5 expression is disrupted in a strip of the mandibular arch mesenchyme of mutant embryos (B). C–D. Expression of the intermediate domain marker Nkx3.2 is also disrupted in the mandibular arch mesenchyme of mutant embryos (D). E–F. Double ISH for Dlx5 (magenta) and Nkx3.2 (blue) shows that in the mutant embryos, the residual Nkx3.2 mesenchyme expression (arrow in F) is confined to the domain in which Dlx5 is absent. G–H. Dlx3 expression is absent in the mandibular arch mesenchyme of mutant embryos. I–J. Double ISH for Dlx3 and Nkx3.2 shows that Nkx3.2 mesenchyme expression (blue) is contained within the Dlx3 mesenchyme domain (magenta) in control embryos (I). This overlapping expression is absent in E10.5 Edn1fl/fl;Foxg1-Cre embryos (J). 1, mandibular pharyngeal arch; 2, second pharyngeal arch.
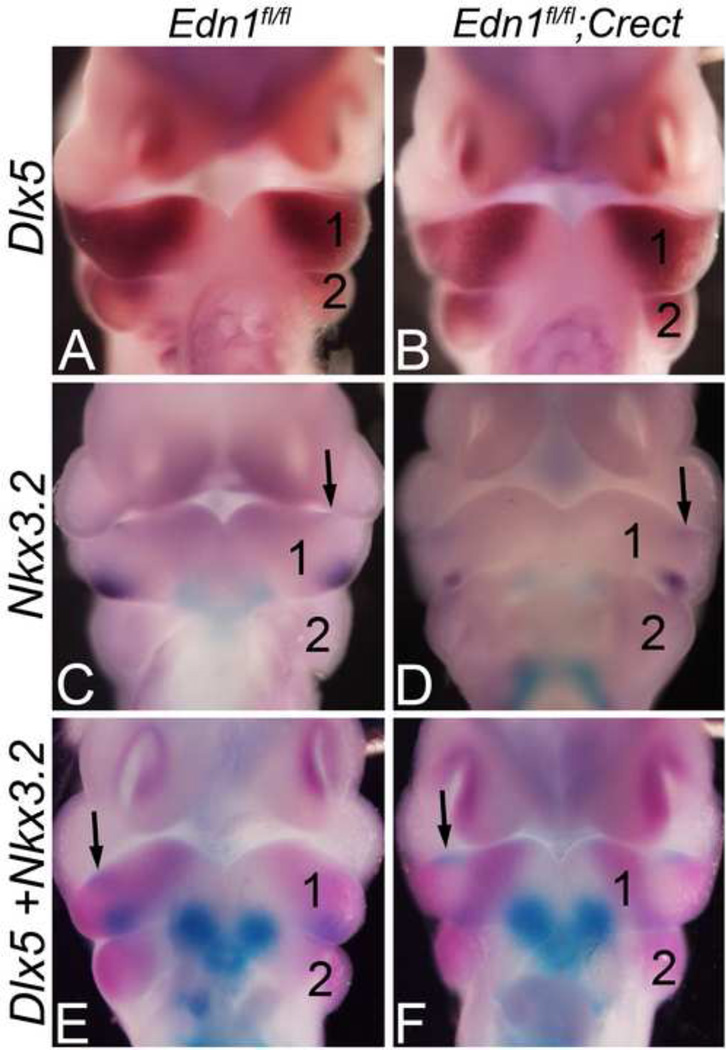
Figure 8. Nkx3.2 expression is disrupted in the intermediate mandibular arch domain of Edn1fl/fl;Crect embryos.
Ventral view of E10.5 Edn1fl/fl;Crect embryos after single (A–D) or double (E–F) ISH for Dlx5 and/or Nkx3.2. A–B. Dlx5 expression is disrupted in a strip of the mandibular arch mesenchyme in Edn1fl/fl;Crect embryos (B). C–D. Mesenchymal expression of Nkx3.2 is disrupted in the mandibular of Edn1fl/fl;Crect embryos, while epithelial expression (arrow) is unaffected (D). E–F. Double ISH for Dlx5 (magenta) and Nkx3.2 (blue) expression. In control embryos, mesenchymal expression of both genes overlaps (E). In Edn1fl/fl;Crect embryos, mesenchymal Nkx3.2 expression is largely absent from the domain in which Dlx5 expression has been lost (F). 1, mandibular pharyngeal arch; 2, second pharyngeal arch.
Like Nkx3.2, Dlx3 is also a marker of the intermediate domain of zebrafish (Talbot et al., 2010). As described above, Dlx3 expression in control embryos is expressed in the proximo-caudal mesenchyme of the mandibular arch and the distal arch ectoderm (Fig. 5B and Fig. 7G), though the region was larger than that of Nkx3.2 (Fig. 7C). In Edn1fl/fl;Foxg1-Cre embryos, the Dlx3 mesenchyme expression was almost completely absent (Fig. 5G and Fig. 7H). To verify that Nkx3.2 and Dlx3 expression domains overlapped in the mouse arch, we again preformed double in situ hybridization analysis. In E10.5 control embryos, Nkx3.2 mesenchyme expression (blue) was indeed contained within the Dlx3 mesenchyme domain (magenta) (Fig. 7I), though the normal mandibular arch expression domain of Dlx3 was difficult to observe due to the overlying Nkx3.2 expression (compare Fig. 7C, G, I). In Edn1fl/fl;Foxg1-Cre embryos, the Dlx5 expression-free domain corresponded to the region in which Nkx3.2 mesenchyme expression was absent and the expression of Dlx3 was severely downregulated (Fig. 7J).
This data suggests that an intermediate domain does exist in the mouse mandibular arch and that Edn1/Ednra signaling is required to establish and/or maintain gene expression in this domain. To prove that this function is due to ectoderm-derived Edn1, we repeated our in situ analysis in E10.5 Edn1fl/fl;Crect embryos. As described above, Dlx5 expression in Edn1fl/fl;Crect embryos was disrupted in a band across the middle portion of the arch (Fig. 8B). This region corresponded to the Nkx3.2 mesenchymal expression domain in control embryos (Fig. 8C) and the region in which Nkx3.2 expression was lost in Edn1fl/fl;Crect embryos (Fig. 8D). This finding was confirmed in Edn1fl/fl;Crect embryos following double in situ hybridization analysis of Nkx3.2/Dlx5 expression, in which expression of Dlx5 and Nkx3.2 were both lost in the same central arch domain. These findings support the idea that the ectoderm is the source of Edn1 critical for establishing/defining the intermediate domain of the mandibular arch.
DISCUSSION
Edn1 is a crucial signaling factor that establishes the D-V patterning of the pharyngeal arches. Here we have demonstrated using conditional gene inactivation that the ectoderm appears to be the required source of Edn1 during mandibular arch patterning and that this requirement appears most important in the intermediate arch domain.
Ectoderm is a required source of Edn1 during patterning of the mandibular first pharyngeal arch
Using Cre/loxP technology, we have taken advantage of mouse genetics to illustrate that the cranial ectoderm is a critical source of Edn1 during facial morphogenesis in mammals. All Cre strains used in this work were able to recombine the R26R allele by E8.5/E9.0, the time period during which Ednra signaling is required for D-V patterning of the mouse pharyngeal arches (Ruest and Clouthier, 2009). We also showed that the conditional Edn1 allele is recombined by E9.0 using all four Cre strains. While not quantitative, the recombination band observed in Edn1fl/fl;Foxa2mcm embryos was by far the weakest. While it is possible that this Cre strain produces a poor recombination efficiency of floxed alleles, we note that Foxa2mcm-mediated recombination of the R26R allele appears robust (Fig. 1P–T and (Park et al., 2008)). More likely, the limited recombination seen using PCR reflects the amount of endoderm contained within the mandibular arch sample used for our assay. Further, because the preproendothelin-1 mRNA has a short intracellular half-life of around 15 minutes (Inoue et al., 1989), we do not believe that mRNA present before gene recombination could significantly contribute to Ednra signaling after Cremediated recombination.
In both Edn1fl/fl;Foxg1-Cre and Edn1fl/fl;Crect embryos, defects in lower jaw structures were present including the presence of duplicated maxillary structures. This was accompanied by a partial loss of Dlx5 and Nkx3.2 in both strains and a loss of Dlx3 in Edn1fl/fl;Foxg1-Cre embryos. Together, our results illustrate that Edn1 from the ectoderm is likely the most crucial source of Edn1 during D-V patterning of the pharyngeal arches. However, a complete loss of mandibular identity as seen in Edn1, Ednra or Dlx5/Dlx6 knockout mice (Beverdam et al., 2002; Depew et al., 2002; Ozeki et al., 2004; Ruest et al., 2004) was not observed even when Edn1 was inactivated in the ectoderm, endoderm and core mesoderm using the Foxg1-Cre strain. One explanation for this partial phenotype is the timing of recombination. The jaw phenotype and related gene expression changes observed in Edn1fl/fl;Foxg1-Cre embryos resemble that of embryos following Ednra antagonism between E8.5 and E9.0 (Ruest and Clouthier, 2009). In this case, normal D-V patterning of the arches would be initiated but then lost, with differential effects observed in the distal and intermediate domains (see below). Another possibility is that the Foxg1-Cre strain did not elicit complete Edn1 inactivation throughout the entire tissue. If conditional gene inactivation was incomplete spatially and/or temporally, residual Edn1 could limit the extent of a jaw phenotype. We also cannot rule out the possibility that endoderm-specific loss of Edn1 could later lead to changes in bone development within the head, as while Edn1fl/fl;Foxa2mcm embryos did not show changes in gene expression, we could not generate E18.5 embryos for analysis of bone and cartilage structures. Likewise, we cannot rule out the possibility that Edn1 from all three arch tissues (ectoderm, mesoderm and endoderm) works in concert to pattern the arches. This could explain the less severe phenotype of Edn1fl/fl;Crect embryos. However, together with the lack of gene expression changes observed in Edn1fl/fl;Myf5-Cre and Edn1fl/fl;Foxa2mcm embryos and the short half live of Edn1 mRNA (Inoue et al., 1989), our data suggest that, as in zebrafish (Miller et al., 2000; Miller et al., 2003; Nair et al., 2007), the Edn1 produced by these layers is not crucial for patterning of the first mandibular arch. This argument is strengthened by recent findings illustrating that while the endoderm is required to achieve normal size and shape of jaw cartilages, it is not required for earlier D-V patterning of the NCC-derived mesenchyme (Balczerski et al., 2012).
Identification of the mouse intermediate domain of the mandibular pharyngeal arch
The most intriguing change observed when Edn1 was inactivated in the ectoderm was loss of Dlx5 expression in a rostrocaudal stripe across the middle of the mandibular arch. We further showed that this domain can be defined by the expression of both Dlx3 and Nkx3.2, downstream mediators of Ednra signaling. This location and expression profile matches the intermediate domain in zebrafish, which plays a crucial role in establishing the zebrafish jaw joint. Loss of zebrafish Edn1 leads to downregulation of nkx3.2 and subsequent fusion of Meckel’s cartilage and the palatoquadrate (Miller et al., 2003). In contrast, loss of Hand2 in both mouse and zebrafish embryos leads to expansion of the Nkx3.2/nkx3.2 domain (Miller et al., 2003) and Tavares and Clouthier, unpublished). While Nkx3.2 and Dlx3 are expressed in the central mandibular arch of the mouse, it has been difficult to truly demarcate this domain and understand its functional significance. Here we have shown that the gene expression domains of both Nkx3.2 and Dlx3 in the mandibular arch corresponds to the central arch domain in which Dlx5 expression is lost, thus marking this affected region as at least a portion of the mammalian intermediate arch domain. Fate mapping of this domain in zebrafish has shown that it gives rise to the dorsal aspects of ventral cartilages and the ventral aspects of dorsal cartilages (Talbot et al., 2010), hence its roll in joint development.
There are obvious differences between the jaw joints of fish and mammals. In zebrafish, Nkx3.2 is required for the proper formation of the jaw joint between Meckel’s cartilage and the palatoquadrate (Miller et al., 2003). In the mouse, this joint appears to correlate with the articulation between the malleus and the incus (Tucker et al., 2004). While this articulation is not disrupted in mouse Nkx3.2−/− embryos, potentially due to evolutionary changes in Gdf5/Gdf6 expression, two genes involved in joint formation, other middle ear structures are abnormal or missing (Tucker et al., 2004). This suggests that Nkx3.2 (and by extension, Dlx3) do indeed mark the intermediate domain of the mammalian mandibular arch. Interestingly, the one common change in the skulls of Edn1fl/fl;Foxg1-Cre and Edn1fl/fl;Crect embryos is a duplication of the jugal bone that occurs concomitant with the loss of malleus and incus, a finding observed in endothelin mouse mutants (Ozeki et al., 2004; Ruest et al., 2004). In control embryos, the malleus and incus articulate through a fibrous joint. Likewise, in mutant embryos, the duplicated jugal bone articulates with the actual jugal bone of the zygomatic arch through a fibrous joint. Thus, while a change in identity of the intermediate domain NCCs has occurred in endothelin mutant embryos, the functional outcome (a joint) is unchanged. This suggests that patterning cues in the arch may occur or be confined by a functional registry that has not been fully appreciated.
The identity of the signals that drives jugal formation in the maxillary prominence is unknown. That a jugal bone forms in the mandibular arch mesenchyme of endothelin mutants could result from the action of mandibular arch signals that normally work with Ednra signaling to drive a different developmental fate. Alternatively, loss of Ednra signaling could result in the inappropriate upregulation of maxillary signals normally involved in jugal determination. The expression of multiple genes is upregulated in mandibular arch in the absence of Ednra signaling, including Dlx2 (Ruest et al., 2004). Interestingly, targeted deletion of Dlx2 leads to disruption of jugal formation (Qiu et al., 1997; Qiu et al., 1995). While we did not observe any gross changes in Dlx2 expression, it is possible that mild changes in the Dlx2 expression boundary sufficient to change the local fate of the mandibular mesenchyme could exist. Further molecular dissection of this region will be required to address these questions.
Sensitivity of the intermediate domain to loss of Ednra signaling
One question that remains from this and previous studies examining variable loss of Ednra signaling in mouse and zebrafish is why this intermediate domain is so sensitive to the level of Edn1. Our gene expression analysis of Edn1fl/fl;Foxg1-Cre embryos illustrates that while distal expression of Dlx5 is present at E9.5, normal proximal/intermediate expression is absent. However, by E10.5, the proximal expression of Dlx5 has recovered while the intermediate expression is still absent. In addition, we have previously shown that following short-term pharmacological antagonism of Ednra signaling in mouse embryos, gene expression in this domain is the first and longest affected (Ruest and Clouthier, 2009). Taken together, the most likely explanations is that Edn1 initiates patterning in both the distal and intermediate domains, but recombination via the Cre strains we have used occurs after Edn1/Ednra signaling has initiated patterning cascades in the distal arch. In this case, we would expect that if Edn1 were inactivated earlier in arch morphogenesis (E8.0–E8.25), we would more closely recapitulate the Edn1/Ednra/Ece1 mutant phenotype (reviewed in (Clouthier et al., 2010). Further, our findings suggest that alternative mechanisms can compensate for loss of Ednra signaling to induce Dlx5 expression in the proximal arch. Together, these findings illustrate that the uniform Dlx5 expression pattern that exists in the mandibular arch of wild type embryos masks a far more complex and overlapping set of control mechanisms. Future advances in Cre transgenic mouse strains that activate earlier or target specific arch sub-domains should make these hypotheses testable.
The hypothesis presented above requires the action of one or more additional factors to maintain Edn1-induced signaling pathways within the distal mandibular arch following loss of Edn1. Our results clearly show that such a molecule would have to be distal-specific, since intermediate gene expression is neither initiated nor maintained. One candidate for this molecule could be a Bmp, as Bmp signaling in zebrafish appears to pattern the distal arch by inducing edn1 expression and later maintaining Edn1-induced expression of hand2 (Alexander et al., 2011). In addition, overexpression of Bmp4 in mice leads to upregulation of Hand2 expression independent of Dlx5/Dlx6 (Bonilla-Claudio et al., 2012). However, if a BMP-dependent pathway is involved in the mouse model, then Bmp2 and/or Bmp7 would be the most likely candidates, as ectodermal Bmp4 expression is lost in Ednra−/− mutants (Ruest et al., 2004). Another factor that works in concert with Ednra signaling to induce Dlx5/Dlx6 expression during mandibular arch development is Mef2c (Arnold et al., 2007; Miller et al., 2007). However, mef2ca mutant zebrafish embryos have jaw joint defects, suggesting that its function is not restricted to the distal/ventral arch (Miller et al., 2007). Closer examination of other distal/ventral ectodermal signals and their relationship with Ednra signaling may shed additional light on this question.
While less likely, another possibility is that the increased sensitivity of the intermediate domain to loss of Ednra signaling is due to differential competence of NCC populations within the arch to respond to Edn1. Whether this actually occurs is not clear, as Ednra signaling in the mouse is required in a cell autonomous manner in both distal and intermediate domain derivatives (Clouthier et al., 2003). However, extrapolation of findings from NCC fate mapping in the chick suggest that the mouse mandible (a distal arch derivative (Ruest et al., 2003)) is composed almost solely of posterior midbrain-derived NCCs, while more proximal structures, including the tympanic ring and gonial bones, also contain NCCs from rhombomeres 1 and 2 (Kontges and Lumsden, 1996). It is plausible that while midbrain versus r1/r2 NCC populations could be equally dependent on Ednra signaling, r1/r2 cells would be less competent to respond to the signal that rescues gene expression in the distal (midbrain-derived) arch in endothelin mutants. It is also possible that both populations are equally competent to respond to additional signals, but mechanisms exist to limit expression of the compensating factor to the distal domain. Detailed analysis of Ednra signaling in these specific NCC populations will be required to answer these questions.
Supplementary Material
HIGHLIGHTS.
The source of endothelin-1 (Edn1) during pharyngeal arch morphogenesis is examined.
Only loss of Edn1 from the ectoderm leads to defects in facial development.
Conditional gene deletion of Edn1 allowed visualization of the intermediate domain.
Intermediate arch gene expression was most sensitive to loss of Edn1.
Loss of intermediate arch gene expression correlated with proximal jaw defects.
ACKNOWLEDGEMENTS
We would like to thank Alicia Navetta for technical assistance and James F. Martin and Anne M. Moon for the tamoxifen-inducible Foxa2mcm mice. This work was supported by NIH/NIDCR research grant DE014181 (to D.E.C.). The NIH had no involvement in the design, data collection and interpretation or the decision to submit this article for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe M, Ruest L-B, Clouthier DE. Fate of cranial neural crest cells during craniofacial development in endothelin-A receptor deficient mice. Int. J. Dev. Biol. 2007;51:97–105. doi: 10.1387/ijdb.062237ma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C, Zuniga E, Blitz IL, Wada N, LePabic P, Javidan Y, Zhang T, Cho KW, Crump JG, Schilling TF. Combinatorial roles for Bmps and Endothelin 1 in patterning the dorsal-ventral axis of the craniofacial skeleton. Development. 2011;138:5135–5146. doi: 10.1242/dev.067801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev. Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Balczerski B, Matstani M, Castillo P, Osborne N, Stainier DY, Crump JG. Analysis of sphingosine-1-phosphate signaling mutants reveals endodermal requirements for the growth but not the dorsoventral patterning of jaw skeletal precursors. Dev. Biol. 2012;362:230–241. doi: 10.1016/j.ydbio.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron F, Woods C, Kuhn K, Bishop J, Howard MJ, Clouthier DE. Downregulation of Dlx5 and Dlx6 expression by Hand2 is essential for initiation of tongue morphogenesis. Development. 2011;138:2249–2259. doi: 10.1242/dev.056929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverdam A, Merlo GR, Paleari L, Mantero S, Genova F, Barbieri O, Janvier P, Levi G. Jaw transformation with gain of symmetry after Dlx5/Dlx 6 inactivation: mirror of the past. Genesis. 2002;34:221–227. doi: 10.1002/gene.10156. [DOI] [PubMed] [Google Scholar]
- Bonilla-Claudio M, Wang J, Bai Y, Klysik E, Selever J, Martin JF. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development. 2012;139:709–719. doi: 10.1242/dev.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner-Fraser M. Origins and developmental potential of the neural crest. Exp. Cell Res. 1995;218:405–417. doi: 10.1006/excr.1995.1173. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson J, R E. Recent advances in craniofacial morphogenesis. Dev. Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Garcia E, Schilling TF. Regulation of facial morphogenesis by endothelin signaling: insights from mouse and fish. Am. J. Med. Genet. A. 2010;152A:2962–2973. doi: 10.1002/ajmg.a.33568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Hosoda K, Richardson JA, Williams SC, Yanagisawa H, Kuwaki T, Kumada M, Hammer RE, Yanagisawa M. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Hammer RE, Richardson JA, Yanagisawa M. Cell-autonomous and nonautonomous actions of endothelin-A receptor signaling in craniofacial and cardiovascular development. Dev. Biol. 2003;261:506–519. doi: 10.1016/s0012-1606(03)00128-3. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. 2002;298:381–385. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/Inactivation in the mouse. Dev. Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Inoue A, Yanagisawa M, Takuwa Y, Mitsui Y, Kobayashi M, Masaki T. The human preproendothelin-1 gene. J. Biol. Chem. 1989;264:14954–14959. [PubMed] [Google Scholar]
- Kimmel CB, Ullmann B, Walker M, Miller CT, Crump JG. Endothelin 1-mediated regulation of pharyngeal bone development in zebrafish. Development. 2003;130:1339–1351. doi: 10.1242/dev.00338. [DOI] [PubMed] [Google Scholar]
- Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, Oda H, Kuwaki T, Cao W-H, Kamada N, Jishage K, Ouchi Y, Azuma S, Toyoda Y, Ishikawa T, Kumada M, Yazaki Y. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM. The neural crest. New York: Cambridge Univ Press; 1982. [Google Scholar]
- Le Douarin NM, Ziller C, Couly GF. Patterning of neural crest derivatives in the avian embryo: in vivo and in vitro studies. Dev. Biol. 1993;159:24–49. doi: 10.1006/dbio.1993.1219. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Sprawson N, Graham A. Segmental origin and migration of neural crest cells in the hindbrain region of the chick embryo. Development. 1991;113:1281–1291. doi: 10.1242/dev.113.4.1281. [DOI] [PubMed] [Google Scholar]
- Maemura K, Kurihara H, Kurihara Y, Oda H, Ishikawa T, Copeland NG, Gilbert DJ, Jenkins NA, Yazaki Y. Sequence analysis, chromosomal location, and developmental expression of the mouse preproendothelin-1 gene. Genomics. 1996;31:177–184. doi: 10.1006/geno.1996.0029. [DOI] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K-H, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3838. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Miller CT, Swartz ME, Khuu PA, Walker MB, Eberhart JK, Kimmel CB. mef2ca is required in cranial neural crest to effect Endothelin1 signaling in zebrafish. Dev. Biol. 2007;308:144–157. doi: 10.1016/j.ydbio.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stainier DY, Kimmel CB. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development. 2003;130:1353–1365. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- Nair S, Li W, Cornell R, Schilling TF. Requirements for endothelin type-A receptors and endothelin-1 signaling in the facial ectoderm for the patterning of skeletogenic neural crest cells in zebrafish. Development. 2007;134:335–345. doi: 10.1242/dev.02704. [DOI] [PubMed] [Google Scholar]
- Ozeki H, Kurihara Y, Tonami K, Watatani S, Kurihara H. Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mech. Dev. 2004;121:387–395. doi: 10.1016/j.mod.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Park EJ, Sun X, Nichol P, Saijoh Y, Martin JF, Moon AM. System for tamoxifen-inducible expression of cre-recombinase from the Foxa2 locus in mice. Dev. Dyn. 2008;237:447–453. doi: 10.1002/dvdy.21415. [DOI] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Ghattas I, Meneses JJ, Christensen L, Sharpe PT, Presley R, Pedersen RA, Rubenstein JLR. Role of the Dlx homeobox genes in proximodistal patterning of the branchial arches: Mutations of Dlx-1, Dlx-2, and Dlx-1 and -2 alter morphogenesis of proximal skeletal and soft tissue structures derived from the first and second arches. Dev. Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]
- Qiu M, Bulfone A, Martinez S, Meneses JJ, Shimamura K, Pedersen RA, Rubenstein JLR. Null mutation of Dlx-2 results in abnormal morphogenesis of proximal first and second branchial arch derivatives and abnormal differentiation in the forebrain. Genes Dev. 1995;9:2523–2538. doi: 10.1101/gad.9.20.2523. [DOI] [PubMed] [Google Scholar]
- Ruest L-B, Dager M, Yanagisawa H, Charité J, Hammer RE, Olson EN, Yanagisawa M, Clouthier DE. dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev. Biol. 2003;257:263–277. doi: 10.1016/s0012-1606(03)00068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Clouthier DE. Elucidating timing and function of endothelin-A receptor signaling during craniofacial development using neural crest cell-specific gene deletion and receptor antagonism. Dev. Biol. 2009;328:94–108. doi: 10.1016/j.ydbio.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Xiang X, Lim KC, Levi G, Clouthier DE. Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development. 2004;131:4413–4423. doi: 10.1242/dev.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Kurihara Y, Asai R, Kawamura Y, Tonami K, Uchijima Y, Heude E, Ekker M, Levi G, Kurihara H. An endothelin-1 switch specifies maxillomandibular identity. Proc. Natl. Acad. Sci. U S A. 2008;105:18806–18811. doi: 10.1073/pnas.0807345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Gen. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Talbot JC, Johnson SL, Kimmel CB. hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development. 2010;137:2507–2517. doi: 10.1242/dev.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Weismann KE, Hellstrom M, Soriano P. Early myotome specification regulates PDFGA expression and axial skeleton development. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Watson RP, Lettice LA, Yamada G, Hill RE. Bapx1 regulates patterning in the middle ear: altered regulatory role in the transition from the proximal jaw during vertebrate evolution. Development. 2004;131:1235–1245. doi: 10.1242/dev.01017. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Hammer RE, Richardson JA, Williams SC, Clouthier DE, Yanagisawa M. Role of Endothelin-1/Endothelin-A receptor-mediated signaling pathway in the aortic arch patterning in mice. J. Clin. Invest. 1998a;102:22–33. doi: 10.1172/JCI2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, Clouthier DE, de Wit D, Emoto N, Hammer RE. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998b;125:825–836. doi: 10.1242/dev.125.5.825. [DOI] [PubMed] [Google Scholar]
- Zuniga E, Stellabotte F, Crump JG. Jagged-Notch signaling ensures dorsal skeletal identity in the vertebrate face. Development. 2010;137:1843–1852. doi: 10.1242/dev.049056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.