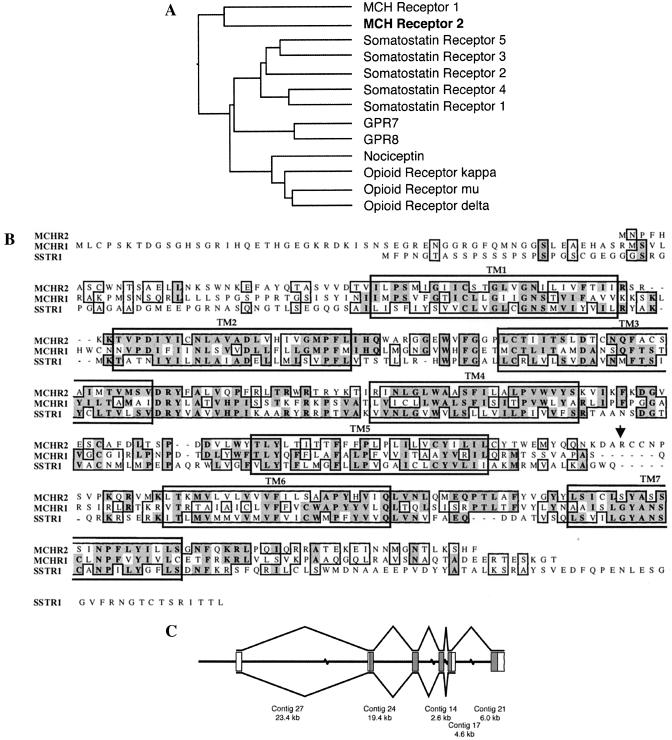
Figure 1.
Analysis of the sequences, genomic structure, and splicing of the MCHR2 gene. (A) Sequence similarity between MCHR2 and related GPCRs. The protein sequence of MCHR2 was aligned with the GPCRs most similar to it, including all of the members of the somatostatin and opioid receptor families and two orphan GPCRs, GPR7 and GPR8. The GCG programs distances and growtree were used to generate a similarity tree based on the alignment. The lengths of the horizontal line segments connecting two GPCRs are inversely proportional to the similarities between their protein sequences. (B) Alignment of the protein sequences of MCHR1, MCHR2, and SSTR1. Identical and similar amino acid residues are boxed, with identical ones in shadowed boxes. The large boxes indicate predicted transmembrane domains. The arrow indicates the end of the putative truncated form of the MCHR2 protein as a result of alternative splicing. (C) MCHR2 exon structure. The MCHR2 mRNA sequence was used to order and orient five of the unordered contigs in the GenBank BAC sequence AC027643. The central line represents the BAC sequence, with discontinuities representing gaps of unknown length between contigs. Boxes represent regions of the genomic sequence containing MCRH2 exons; filled boxes are translated, and open boxes contain 5′ and 3′ untranslated region sequences. Lines above and below the exons indicate the two splicing patterns observed. The position of the 3′ end of the 3′ untranslated region of the longer transcript is unknown. The exon lengths are drawn out of scale with the contig lengths.