Figure 5.
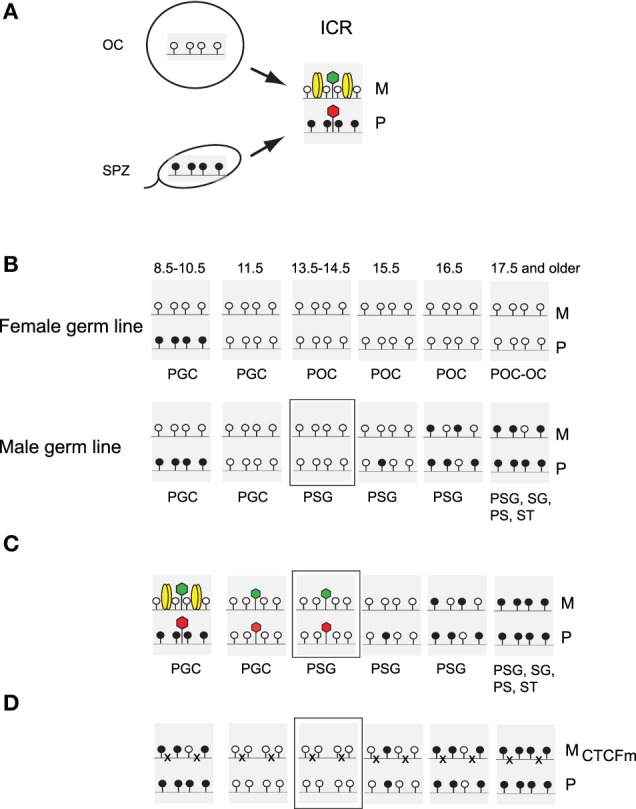
CTCF binding delays de novo methylation of the maternal allele in male germ cells. (A) Differential methylation of the ICR is inherited from the gametes: methylation of the paternal allele (P) from spermatozoa (SPZ) and unmethylation of the maternal allele (M) from oocytes (OC). This primary methylation difference determines CTCF binding and chromatin composition in the soma and likely also in primordial germ cells (PGC), which exhibit imprinted H19 and Igf2 expression. Active or repressive chromatin (green or red hexagon) is present at respective alleles of the ICR. (B) Fate of the imprint in the female and male germ lines. Methylation status of the ICR is depicted in the primordial germ cells (PGC), primary oocytes (POC) and in prospermatogonia (PSG), spermatogonia (SG) pachytene spermatocytes (PS) and round spermatids (ST) with gestational stages in dpc. The developmental stage that appears epigenetically different without DNA methylation is marked with a rectangle. (C) Imprint establishment of the ICR in the normal male germ line. Expected CTCF binding and chromatin composition is depicted in primordial germ cells (PGC). Observed chromatin bias is depicted in prospermatogonia (PSG). Chromatin bias is observed in the normal ICR between the parental alleles in the absence of CpG methylation at 13.5–14.5 dpc. (D) Functional CTCF sites are required for chromatin bias and delayed methylation of the maternally inherited ICR allele. Maternal inheritance of the CTCF binding site mutations abolishes CTCF binding in the maternal allele in PGCs. No chromatin bias is observed between parental alleles at 13.5–14.5 dpc and the maternal allele's methylation is not delayed at 15.5–17.5 dpc.
