TO THE EDITOR
Melanoma is among the most severe and lethal forms of human skin cancer (Mackie, 2006; Miller and Mihm Jr, 2006; Fecher et al., 2007; Gray-Schopfer et al., 2007). For 2008, more than 110,000 new cases of melanoma (about 50,000 melanoma in situ (MIS) and 60,000 invasive melanomas) were estimated with more than 8,000 deaths (American Academy of Dermatology, 2008; Ries et al., 2008). To examine genetic diversity, we studied metastatic melanoma lesions from a patient with xeroderma pigmentosum (XP). XP is a rare genetic disease with defective DNA repair and a more than 1000-fold increase in melanoma frequency (Kraemer et al., 1987, 1994; Ruenger et al., 2008).
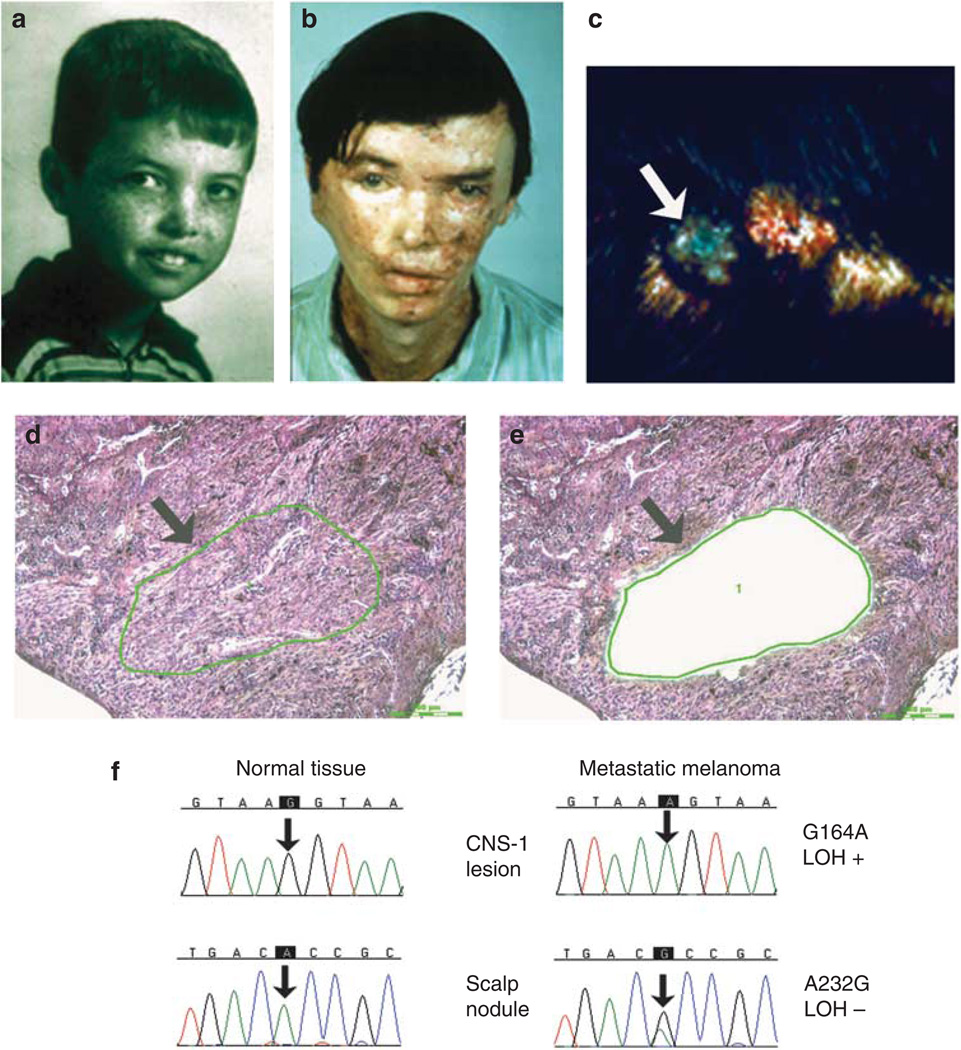
The XP patient, (XP4BE), had no blistering reaction to sunlight despite living on a farm with unrestricted exposure to sunlight. By 8 years of age he had extensive freckling on his face (Figure 1a). At the age of 9 years a large warty tumor was excised from his nose. He subsequently developed more than 100 cancers in sun-exposed skin, primarily basal cell carcinomas and squamous cell carcinomas, which were treated with electrodessication and curettage or surgical excision followed by skin grafting (Figure 1b). At the age of 23 years a primary malignant melanoma of the right post-auricular area had been widely excised, and evidence of spread was found in two of the 35 posterior cervical nodes.
Figure 1. Clinical appearance, histology, and DNA sequencing of metastatic melanoma lesions in patient XP4BE.
(a, b) Comparison of the face of patient XP4BE at the ages of 8 and 27 years. By the age of 27, he had had surgery with grafting for many skin cancers (reproduced from Robbins et al. (1974)). (c) Metastatic melanoma of the scalp before treatment with bis-choroethyl-nitrosourea (arrow). (d) Histology of metastatic melanoma lesion in scalp. Atypical melanocytes are arranged in sheets and nests (arrow) (hematoxylin and eosin (H&E) staining, bar = 200 µm). (e) After capture of the melanoma cells, the remaining tissue was inspected (arrow), and the transfer efficiency of about 500 captured cells was evaluated (H&E staining, bar = 200 µm). (f) Sequencing chromatograms for determination of PTEN mutations. The metastatic melanoma DNA shows a mutation compared with the normal tissue (arrow). LOH, loss of heterozygosity. (Details of method are described in Wang et al. (2009)).
XP4BE was admitted to the NIH Clinical Center in 1969 at the age of 25 years (Robbins et al., 1974) and studied in accordance with the NIH human research guidelines then in effect. He was the first XP “variant” patient recognized, a form of XP with normal nucleotide excision repair (Robbins et al., 1974). His cells were subsequently found to have a defect in the error-prone polymerase, polymerase eta (Johnson et al., 1999; Masutani et al., 1999). At admission, his neurological examination was normal. In 1971, a melanoma nodule was present on his scalp (Figure 1c). He had no beneficial response to two courses of bis-choroethyl-nitrosourea (BCNU), steroids, or radiation to the brain and lumbar areas for treatment of the metastatic melanoma. He died at the age of 27 years in 1971. Autopsy performed at NIH revealed thromboemboli with pulmonary infarctions, aspergillus pneumonia and cerebral abscess, and metastatic melanoma involving his brain, spinal cord, stomach, small bowel, liver, gall bladder, adrenal glands, kidneys, lymph nodes, left testis, right lung, pancreas, thyroid gland, and soft tissue of the right thigh. The paraffin blocks from this autopsy were recently retrieved, sectioned, and stained. We do not have tissue from the primary lesion, which was removed before coming to NIH. We analyzed seven metastatic lesions from this patient, the metastatic scalp nodule obtained before BCNU treatment (Figure 1c–e), and six lesions obtained at autopsy (central nervous system (CNS) × 2, muscle, liver, kidney, and pancreas) (Table 1) as well as normal tissue. Using laser capture microdissection, we isolated about 500 tumor or normal cells for DNA analysis (Figure 1d and e) as described previously (Wang et al., 2009). To minimize formalin sequencing artifacts (Williams et al., 1999), we used more than 500 cells for analysis and repeated the sequencing on independent PCR products obtained from adjacent regions of the tumor for several tumors.
Table 1.
Mutations in PTEN, NRAS, and BRAF in metastatic melanoma lesions from patient XP4BE
| Metastatic lesion no. |
Metastatic melanoma site |
PTEN | NRAS | BRAF | ||||
|---|---|---|---|---|---|---|---|---|
| Mutation | UV type mutation?1 |
LOH?2 | Amino-acid change |
Mutation | Amino-acid change |
Mutation | ||
| 1 | Liver | gGc131gAc3 | + | − | Gly44Asp3 | Normal | Normal | Normal |
| 2 | CNS-1 | aGg164aAg3 | + | + | Arg55Lys3 | NT4 | NT | Normal |
| 2 | CNS-1 | tAt521tGt | − | − | Tyr174Cys | |||
| 3 | Skeletal muscle | tCa733tTa | + | − | Gln245X | NT | NT | NT |
| 3 | Skeletal muscle | gTc882gCc | + | − | Ser294Ser5 | |||
| 4 | Kidney | aGg164aAg3 | + | − | Arg55Lys3 | aTc99aCc | Asp33Asp5 | NT |
| 4 | Kidney | aAc499aGc | + | − | Thr167Ala6 | |||
| 4 | Kidney | aGg663aTg | + | + | Lys221Asn | |||
| 5 | Pancreas | tAa162tGa | + | − | Val54Val5 | aAt93aGt | Glu31Glu5 | Normal |
| 5 | Pancreas | gGa494gTa | + | − | Gly165Val | cCa105cTa | Thr35Thr5 | |
| 5 | Pancreas | tTa562tCa | + | − | Tyr188His | |||
| 6 | Scalp | gAt152gGt | + | − | Asp51Gly | NT | NT | Normal |
| 6 | Scalp | cAc232cGc | − | − | Thr78Ala | |||
| 6 | Scalp | aAg548aGg | + | − | Lys183Arg | |||
| 6 | Scalp | gTt583gCt | + | − | Phe195Leu | |||
| 7 | CNS-2 | Normal | Normal | Normal | Normal | Normal | ||
+, Dipyrimidine; −, not dypyrimidine.
+, Loss of heterozygosity; −, no loss of heterozygosity.
Mutation reported in in situ melanoma in XP patients (Wang et al., 2009).
NT, not tested.
Synonymous mutation (no amino-acid change).
Mutation reported in an astrocytoma (Raffel et al., 1999).
We studied the tumor suppressor gene PTEN (phosphatase and tensin homolog), which is one of the most frequently mutated genes in human cancer, including melanoma (Goel et al., 2006; Baker, 2007) as well as the NRAS and BRAF oncogenes that have been reported to be mutated in melanomas (Curtin et al., 2005). We used sequencing techniques previously described (Curtin et al., 2005; Wang et al., 2009 and references therein). Previously, we screened 59 primary cutaneous melanomas from 8 other XP patients and 56% had PTEN base substitution mutations. There were 1–4 mutations in individual melanomas, including MIS, the earliest stage of melanoma (Wang et al., 2009).
Table 1 summarizes the pathological and mutational features of all seven metastatic melanoma lesions tested. Samples from six (86%) of the melanomas showed base substitution mutations in the PTEN tumor suppressor gene. No insertions or deletions were observed. Fifteen base substitution mutations were detected and 14 of these were different from each other. There was 1 nonsense mutation (Gln245X), 12 missense mutations, and 2 synonymous mutations that did not alter the amino-acid sequence. Individual metastatic lesions showed marked genetic diversity with 1–4 different PTEN base substitution mutations: the liver lesion had one mutation; a CNS and a skeletal muscle metastasis each had two mutations; kidney and pancreas metastases each had three mutations; and the metastatic melanoma lesion on the scalp had four different mutations. Thirteen (87%) of the mutations were present at dipyrimidine sites, a feature of UV-induced mutations (Wang et al., 2009). Loss of heterozygosity was found in two of the metastatic melanomas that also had PTEN missense mutations. A Thr167Ala missense mutation was reported previously in an astrocytoma (Raffel et al., 1999). The missense mutations were located in the dual-specificity protein phosphatase domain (amino acids 25–179) as well as in the calcium/lipid binding region (amino acids 190–347).
Two of four (50%) of the metastatic lesions tested had NRAS base substitution mutations. One mutation in NRAS was found in the kidney metastasis and two NRAS mutations were found in the pancreas metastasis. However, these three NRAS mutations were all synonymous. None of the five metastatic lesions tested had BRAF base substitution mutations. The low frequency of mutations in NRAS and BRAF contrasts with the higher frequency of mutations in the PTEN gene obtained from the same samples and suggests that PTEN mutations were not the result of formalin artifacts that would be expected to similarly affect all genes (Williams et al., 1999). Thus, mutations in the NRAS and BRAF oncogenes did not appear to have a major role in metastasis of melanomas in this XP patient.
Patient XP4BE presented with metastatic lesions. In our study of early melanomas in other XP patients (Wang et al., 2009), we found the UV type mutations, gGc131gAc and aGg164aAg, in MIS lesions. These same mutations were also present in three of the metastatic lesions (liver, CNS, and kidney) (Table 1) and might have been present in the primary melanoma(s) in XP4BE. These mutations result in amino-acid alterations (Gly44Asp and Arg55Lys) in the protein phosphatase domain of PTEN and may represent persistent UV-induced genetic alterations associated with metastasis.
Surprisingly, there was no single mutation present in all of the metastatic lesions, as would be expected if all of the metastases arose from a single primary melanoma (Wang et al., 2006; Sabatino et al., 2008). Multiple tumor samples from the same patient had different mutations, indicating the presence of different clonal populations of tumor cells in different metastatic lesions. Independent PCR amplification of laser capture microdissected tumor tissue from adjacent areas of tumors 1, 2, and 4 revealed the identical mutations in PTEN exon 2 as shown in Table 1, with no new mutations found, which is evidence that these mutations were not formalin artifacts (Williams et al., 1999). In addition, normal PTEN sequence was obtained for exons 4, 6, 7, and 8 for tumors 1–6, suggesting that there were local sequence variations in these metastatic melanomas. These metastatic lesions with different PTEN mutations might have arisen from different primary melanomas. The high frequency of UV type mutations in these metastatic lesions (87 vs 54% expected (Wang et al., 2009)) is consistent with their origin from sunlight-induced primary melanomas. Alternatively, alterations in other gene(s) might have induced the metastases (Wang et al., 2006; Sabatino et al., 2008). As in the primary melanomas (Wang et al., 2009), the finding of multiple PTEN mutations in metastatic lesions indicates that there is a marked genetic diversity in these tumors, perhaps reflecting the hypermutability of XP variant cells (Waters et al., 1993; Stary et al., 2003; Wang et al., 2007). A similar genetic heterogeneity of metastatic melanomas from non-XP patients was reported based on the studies of loss of heterozygosity and X-chromosome inactivation (Katona et al., 2007). This genetic diversity may affect melanoma development, progression toward metastasis, and response to therapy.
ACKNOWLEDGMENTS
We thank Dr Jay Robbins, formerly of the NCI Dermatology Branch, for the initial ascertainment and DNA repair assessment of this patient and Ms Silke Williams, NCI Pathology Branch archivist, for her excellent assistance in obtaining the autopsy materials from 1971. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
REFERENCES
- American Academy of Dermatology. Melanoma Fact Sheet. 2008 http://www.aad.org/media/background/factsheets/fact_melanoma.html. [Google Scholar]
- Baker SJ. PTEN enters the nuclear age. Cell. 2007;128:25–28. doi: 10.1016/j.cell.2006.12.023. [DOI] [PubMed] [Google Scholar]
- Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- Fecher LA, Cummings SD, Keefe MJ, et al. Toward a molecular classification of melanoma. J Clin Oncol. 2007;25:1606–1620. doi: 10.1200/JCO.2006.06.0442. [DOI] [PubMed] [Google Scholar]
- Goel VK, Lazar AJ, Warneke CL, et al. Examination of mutations in BRAF, NRAS, and PTEN in primary cutaneous melanoma. J Invest Dermatol. 2006;126:154–160. doi: 10.1038/sj.jid.5700026. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, et al. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- Katona TM, Jones TD, Wang M, et al. Genetically heterogeneous and clonally unrelated metastases may arise in patients with cutaneous melanoma. Am J Surg Pathol. 2007;31:1029–1037. doi: 10.1097/PAS.0b013e31802b3488. [DOI] [PubMed] [Google Scholar]
- Kraemer KH, Lee MM, Andrews AD, et al. The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer. The xeroderma pigmentosum paradigm. Arch Dermatol. 1994;130:1018–1021. [PubMed] [Google Scholar]
- Kraemer KH, Lee MM, Scotto J. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch Dermatol. 1987;123:241–250. doi: 10.1001/archderm.123.2.241. [DOI] [PubMed] [Google Scholar]
- Mackie RM. Long-term health risk to the skin of ultraviolet radiation. Prog Biophys Mol Biol. 2006;92:92–96. doi: 10.1016/j.pbiomolbio.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- Raffel C, Frederick L, O’Fallon JR, et al. Analysis of oncogene and tumor suppressor gene alterations in pediatric malignant astrocytomas reveals reduced survival for patients with PTEN mutations. Clin Cancer Res. 1999;5:4085–4090. [PubMed] [Google Scholar]
- Ries LAG, Melbert D, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2005. Bethesda, MD: National Cancer Institute; 2008. [Google Scholar]
- Robbins JH, Kraemer KH, Lutzner MA, et al. Xeroderma pigmentosum. An inherited disease with sun sensitivity, multiple cutaneous neoplasms, and abnormal DNA repair. Ann Intern Med. 1974;80:221–248. doi: 10.7326/0003-4819-80-2-221. [DOI] [PubMed] [Google Scholar]
- Ruenger TM, DiGiovanna JJ, Kraemer KH. Hereditary diseases of genome instability and DNA repair. In: Wolff K, et al., editors. Fitzpatrick’s Dermatology in General Medicine. New York: McGraw Hill; 2008. pp. 1311–1325. [Google Scholar]
- Sabatino M, Zhao Y, Voiculescu S, et al. Conservation of genetic alterations in recurrent melanoma supports the melanoma stem cell hypothesis. Cancer Res. 2008;68:122–131. doi: 10.1158/0008-5472.CAN-07-1939. [DOI] [PubMed] [Google Scholar]
- Stary A, Kannouche P, Lehmann AR, et al. Role of DNA polymerase eta in the UV-mutation spectrum in human cells. J Biol Chem. 2003;278:18767–18775. doi: 10.1074/jbc.M211838200. [DOI] [PubMed] [Google Scholar]
- Wang E, Voiculescu S, Le PI, et al. Clonal persistence and evolution during a decade of recurrent melanoma. J Invest Dermatol. 2006;126:1372–1377. doi: 10.1038/sj.jid.5700193. [DOI] [PubMed] [Google Scholar]
- Wang Y, DiGiovanna JJ, Stern JB, et al. Evidence of ultraviolet type mutations in xeroderma pigmentosum melanomas. Proc Natl Acad Sci USA. 2009;106:6279–6284. doi: 10.1073/pnas.0812401106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Woodgate R, McManus TP, et al. Evidence that in xeroderma pigmentosum variant cells, which lack DNA polymerase eta, DNA polymerase iota causes the very high frequency and unique spectrum of UV-induced mutations. Cancer Res. 2007;67:3018–3026. doi: 10.1158/0008-5472.CAN-06-3073. [DOI] [PubMed] [Google Scholar]
- Waters HL, Seetharam S, Seidman MM, et al. Ultraviolet hypermutability of a shuttle vector propagated in xeroderma pigmentosum variant cells. J Invest Dermatol. 1993;101:744–748. doi: 10.1111/1523-1747.ep12371686. [DOI] [PubMed] [Google Scholar]
- Williams C, Ponten F, Moberg C, et al. A high frequency of sequence alterations is due to formalin fixation of archival specimens. Am J Pathol. 1999;155:1467–1471. doi: 10.1016/S0002-9440(10)65461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]