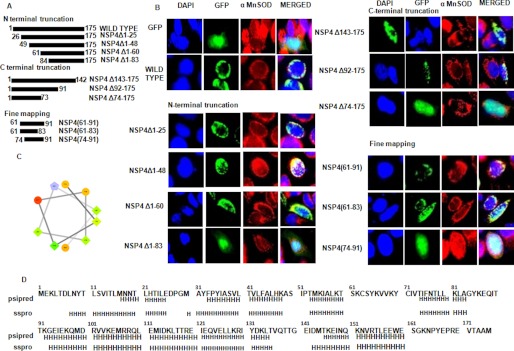
FIGURE 4.
Identification of mitochondrial translocation sequence of NSP4. A, schematic representation of N- and C-terminal NSP4 mutants generated in pACGFP1-c2 vector as described under “Experimental Procedures.” B, 60–83 amino acids is the shortest region of NSP4 that acts as MTS. HeLa cells were transfected with either empty GFP-expressing vector or wild type NSP4 or mutant constructs of NSP4 as GFP fusion protein (green). After 16 h of transfection, cells were fixed and stained with anti-α-MnSOD antibody (mitochondrial marker) followed by rhodamine-labeled (red) secondary antibody and DAPI (blue). The cells were visualized with a fluorescence microscope to assess localization of NSP4 (green) and mitochondria (red). C, helical wheel representation (helical wheel projection; Don Armstrong) of a predicted helix formed by residues 74–83 of NSP4. Hydrophilic residues are shown as circles, hydrophobic residues as diamonds, potentially negatively charged as triangles, and potentially positively charged as pentagons. The most hydrophobic residue is green, and the amount of green is decreasing proportionally to the hydrophobicity, with zero hydrophobicity coded as yellow. Hydrophilic residues are coded red with pure red being the most hydrophilic (uncharged) residue, and the amount of red decreasing proportionally to the hydrophilicity. The potentially charged residues are light blue. D, putative helix prediction (H, helix) within NSP4 sequence using PSIPRED (PSIPRED Server) and sspro (Phyre Server) programs.