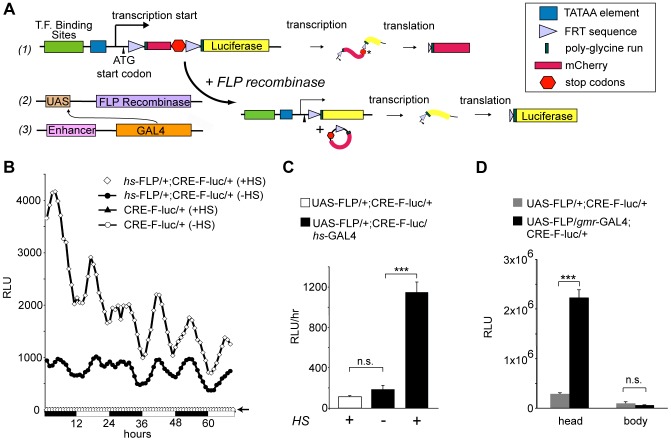
Figure 1. Developing a spatially restricted reporter system.
(A) A cartoon of the reporter system. Three DNA binding sites are placed upstream of the CaSpeR TATAA sequence, followed by transcription (arrow) and translation (arrowhead) initiation sites, an FRT-flanked open reading frame for mCherry (ORF) including two tandem stop codons, and the luciferase ORF (#1). Targeted GAL4 expression (#3) drives the expression of UAS-FLP (#2), which catalyzes site-specific recombination at the FRT sites, activating the reporter. (B) CRE-F-luc reporter activity is FLP-dependent. Singly (CRE-F-luc) or doubly transgeneic flies (hs-FLP/+;CRE-F-luc/+) are maintained under 12∶12 LD conditions. Flies are exposed to heat-shock (+HS) or not (−HS), and measured for in vivo luminescence. The relative luminescence is plotted as a function of time, with daytime (white bars) and nighttime (black bars) durations indicated below the graph. Each data point represents the average hourly luminescence counts in relative light units (RLU) of 24 flies. (C) FLP protein expressed using the GAL4-UAS system can activate the reporter. Reporter activity (Y-axis, the mean hourly relative light units [RLUs] over a 4-day window) is plotted as a function of genotype (indicated with different colored bars) and treatment (+/− HS, heat shock). Error bars = S.E.M. *** p<0.001, Student' T-test, n = 24; n.s. signifies not significant. Similar statistical comparisons are made for the remaining figures. (D) Anatomical specificity of gmrlong-GAL4 driven reporters. In vitro luciferase activity measured in extracts made from isolated heads and bodies. The relative light units (Y-axis) are plotted as a function of the genotype (shown in gray [UAS-FLP/+; CRE-F-luc/+ or black [UAS-FLP/gmrlong-GAL4; CRE-F-luc/+]) or tissue source (head versus body).