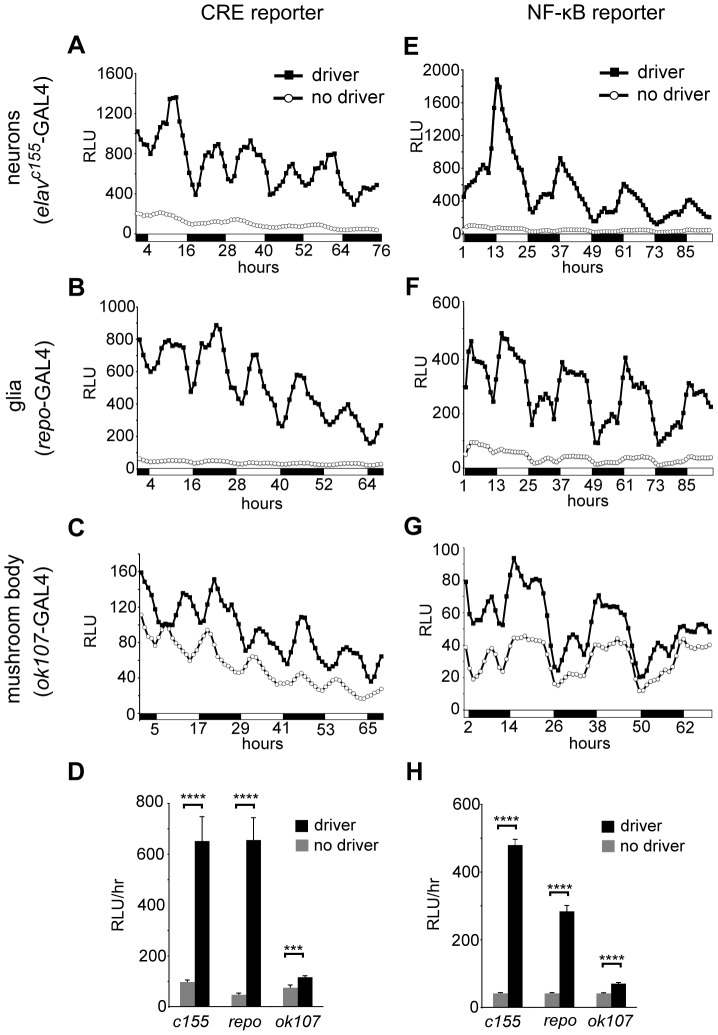
Figure 3. dCREB2 and NF-κB activity cycle in multiple tissues.
Reporter activity is plotted as a function of circadian time. Each different cellular population is described on the left, and the tissue-specific driver used to activate the reporter is shown. In each panel, a comparison is shown between the background (denoted no driver) and activated (driver) expression levels. The activity is shown for the CRE-F-luc (panels A–C) and the NFκB-F-luc (panels E–G) reporters in neurons (panels A and E), glia (panels B and F) and in the mushroom body (panels C and G). The average activity over the duration of the experiment for each driver is quantified for the CRE-F-luc (panel D) and NFκB-F-luc (panel H) reporters. All bars represent mean hourly counts from 24–28 flies. Error bars = S.E.M, ***p<0.001, **** p<0.0001.