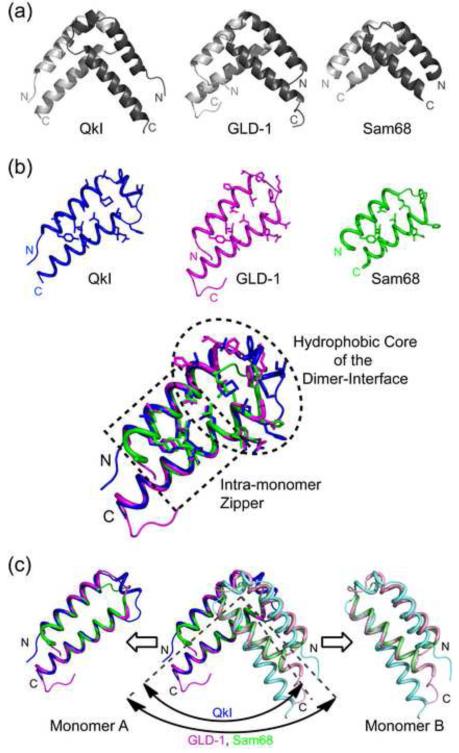
Figure 5. Comparison of the QkI-Qua1 structure (pdb 4DNN) to the GLD-1 (pdb 3K6T) and Sam68 (pdb 2XA6).
(a) Ribbon presentation. QkI-Qua1 features an additional 3 residues in the turn region that enable to form an additional short helix and expand the surface area of the dimer interface. (b) While the two main helices in each monomer overlay precisely for all three Qua1 domains, the two QkI-Qua1 monomers are stacked at a narrower angle compared to the GLD-1 and Sam68 homodimers. (c) Comparison of dimer interface and zipper residues (shown as sticks) of QkI, GLD-1 and Sam68 Qua1 monomers and overlay of the three structures. While the monomer zipper is highly conserved, more variation is found in the hydrophobic dimer interface residues. All three proteins use stacking of conserved Phe residues as the core of the dimer interface, but the exact position of these residues varies to some degree. The additional helix in the QkI structure broadens the dimer interface significantly and allows for more hydrogen bond interactions across the edge of the interface (see also Figure 2).