Figure 1. The Exo9 core alters exoribonuclease, endoribonuclease and RNA binding activities of Rrp44.
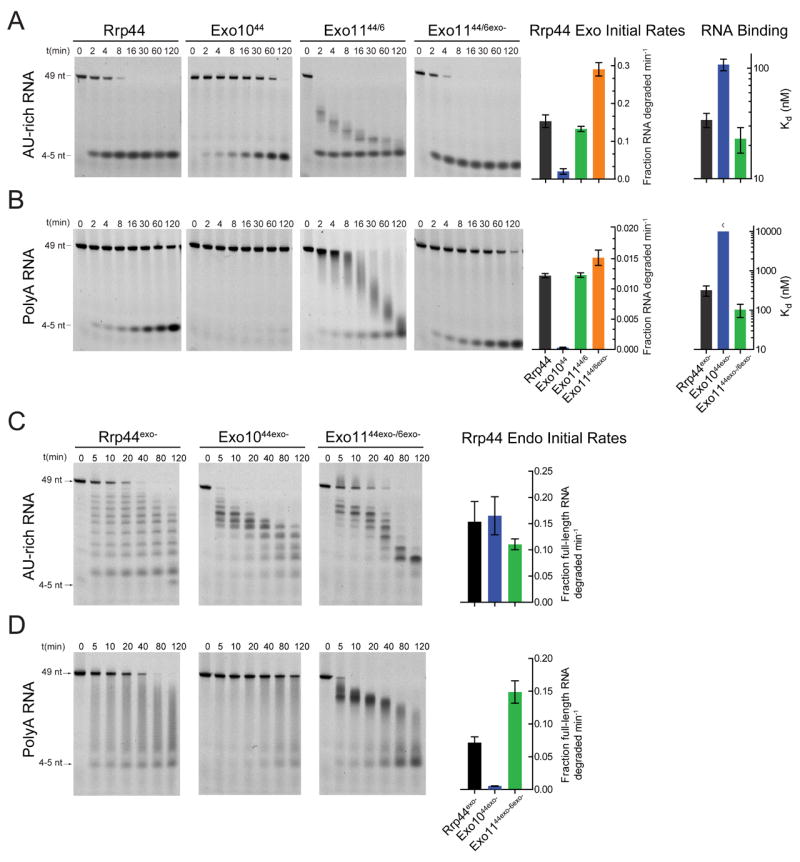
Association of Rrp44 with Exo9 (Exo1044) diminishes the exoribonuclease and RNA binding activities on (A) AU-rich and (B) polyA RNAs. Rrp6 stimulates Rrp44 activity in Exo1144/6 independent of Rrp6 catalytic activity. Multiple turnover RNA decay assays performed using 5’ fluorescein labeled RNAs and reaction products separated by Urea-PAGE. Representative gels and quantitation. Bar graphs depicting initial rates of Rrp44-mediated exoribonuclease activity determined from data obtained in the linear range. Bar graphs depicting dissociation constants (Kd) were derived by fluorescence polarization of catalytically dead variants of free and Exo9 core-associated Rrp44 assayed using 5’ fluorescein labeled (A) AU-rich or (B) polyA RNA. The pattern of intermediates generated by Rrp44exo- changes after association with Exo9 (Exo1044exo-) or Exo9 and Rrp6exo- (Exo1144exo-/6exo-) for (C) AU-rich and (D) polyA RNA (See Figure S3C for assays using unlabeled RNA). The distributive pattern of intermediates observed for Rrp44exo- (short and long 5’ labeled products accumulate simultaneously) is altered to patterns that appear to be generated 3’ to 5’ in Exo1044exo- and Exo1144exo-/6exo- for AU-rich RNA because longer 5’ labeled products appear before appearance of shorter products. Exo1044exo- has weaker endonuclease activity on polyA RNA but is stimulated by addition of Rrp6exo- in Exo1144exo-/6exo-. A stoichiometric ratio of enzymes and 5’-fluorescein RNA (10 nM) was incubated in reaction buffer in the presence of 3 mM MnCl2 (See Experimental Procedures). Error bars represent ± 1 standard deviation as calculated from three independent experiments. Bar graphs color coded according to Table S1.
