Figure 5. Channel occlusion inhibits RNA binding and RNase activities of Rrp44 and Rrp6.
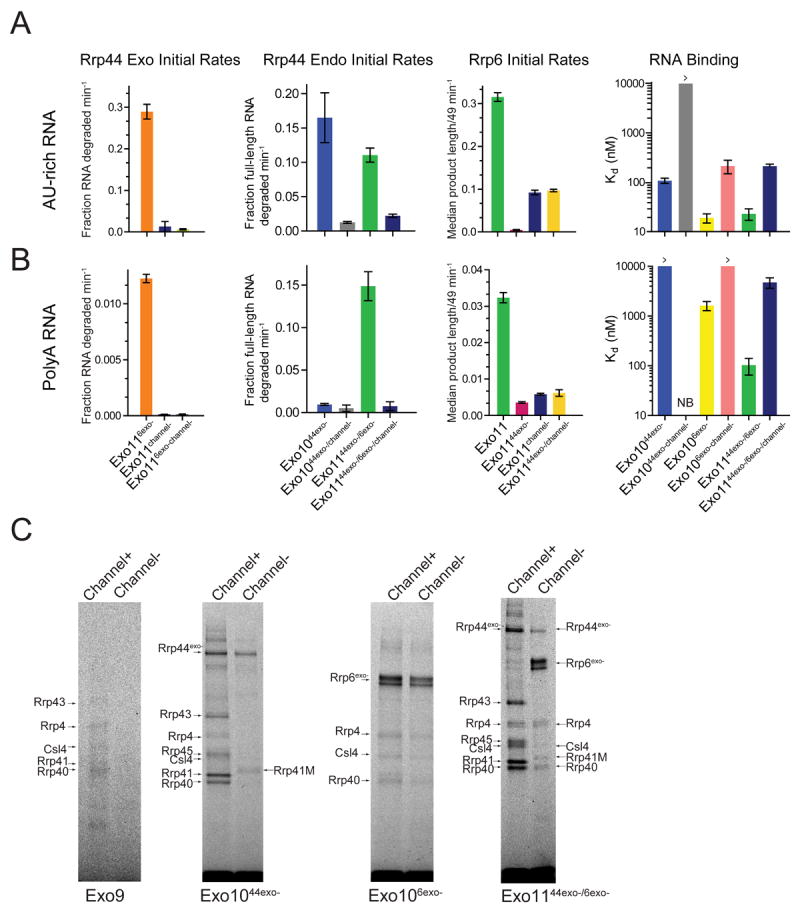
Bar graphs depicting initial rates calculated for Rrp6 and Rrp44 RNase on A) AU-rich and B) polyA RNA. Occluding the central channel of Exo1144/6 (Exo1144/6/channel-) with loop insertions (Rrp41-M/Rrp45-M) inhibits both Rrp44 activities while Exo1144exo-/6/channel- diminishes Rrp6 exoribonuclease activities and alleviates Rrp6 inhibition by Rrp44exo-. Rrp44 endonuclease activities are inhibited by channel occlusion in Exo1044exo- and Exo1144exo-/6exo-. Assays performed in triplicate and initial rates calculated from data obtained in the linear range (Figures S6A and S6C). Bar graphs on the right depict apparent Kd values for various exosome complexes with RNA. Error bars are ± 1 standard deviation. C) Channel occlusion in Exo9 leads to diminished cross-linking (left). Panel to the right shows that channel occlusion in Exo1044exo- leads to loss of most RNA-protein adducts to core subunits. Next panel shows that channel occlusion in Exo106exo- slightly weakens cross-linking to the S1/KH cap proteins and Rrp6. On the right, channel occlusion in Exo1144exo-/6exo- shifts the cross-linking pattern from one involving Rrp44, the PH-like and S1/KH rings to one involving Rrp6 and the S1/KH ring. Products separated by SDS-PAGE and imaged by detecting the fluorescence of the 5’ labeled AU-rich RNA. Major cross-linked species are labeled and indicated by arrows.
