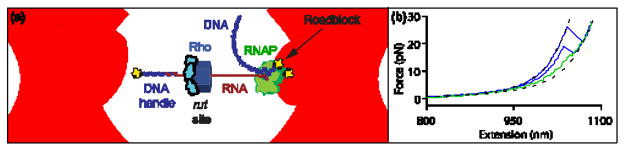
Figure 2. Single-Molecule Dumbbell Assay.
(a) Experimental geometry of the dumbbell assay (not to scale). Tension was applied by moving apart the two trapped beads. The beads (light blue) were maintained in separate optical traps (pink), with a dsDNA handle (dark blue) attached to one and an RNAP molecule (green) to the other by biotin-avidin linkages (yellow). The handle was hybridized to the 5′ end of the RNA transcript (red) via a 25- nt overhang. The RNA was transcribed in situ from a template that carried the rut site of the? tR1 terminator followed by a downstream sequence of variable length (30 bp, 75 bp or 150 bp) ending in a transcriptional roadblock (yellow) that stalls RNAP. The assay permits Rho to bind to the nascent RNA and, in the presence of ATP, translocate towards RNAP.
(b) Representative force-extension curves (FECs). Most FECs (green) resemble those obtained in the absence of Rho and exhibited a small rip due to unfolding of the boxB hairpin in the rut site. FECs displaying larger, high- force rips were observed only in the presence of Rho(blue), and correspond to Rho release from the RNA.