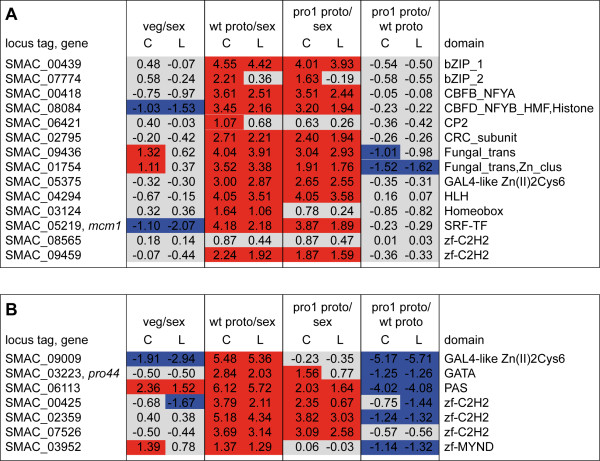
Figure 7.
Expression ratios for transcriptionfactors among the geneswith top 500 readcounts. (A) Expression ratios for the transcription factors among the 500 genes with the highest number of read counts in wild-type and mutant pro1 protoperithecia. Expression of these genes is largely independent of pro1. (B) Expression ratios for the transcription factors among the 500 genes with the highest number of read counts in wild-type protoperithecia but not pro1 protoperithecia. These genes are most likely dependent on pro1 for correct expression. Expression ratios in (A) and (B) are given as log2 values, and log2 ratios >1 and <−1 are indicated in red and blue, respectively. The genes in (A) are mostly not differentially expressed in pro1 protoperithecia compared to wild-type protoperithecia (indicated by the grey coloring), whereas the genes in (B) have a tendency towards down-regulation in pro1 protoperithecia, as expected for genes that are dependent on pro1 for correct expression. Protein domains were predicted with HMMER using the Hidden Markov models from the pfam database [74,75].