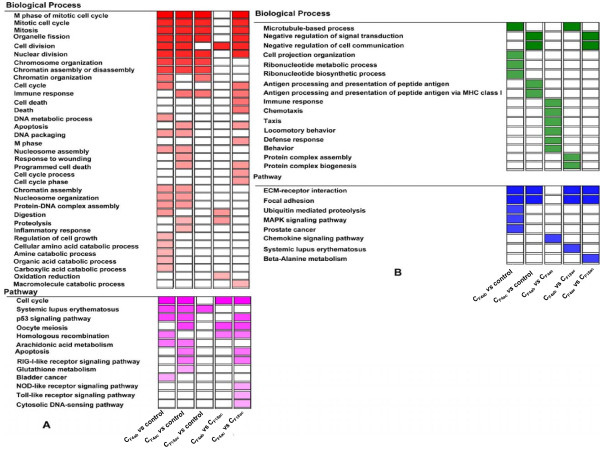
Figure 2.
Gene Ontology and pathway enrichment of differentially expressed genes in all comparisons. CF4ab: IPEC-J2 cells infected with F4ab ETEC; CF4ac: IPEC-J2 cells infected with F4ac ETEC; CF18ac: IPEC-J2 cells infected with F18ac ETEC; Control: the non-infected IPEC-J2 cells. The Unigene ID of the genes constituting every “↑” (up-regulated or more highly expressed after ETEC infection) and “↓” (down-regulated or more lowly expressed after ETEC infection) set in Table1 were subjected to enrichment analysis using DAVID annotation tool. Biological processes and pathways that were significantly enriched (adjusted p-value < 0.05) in at least one cluster were shown in heatmap-like graph. A is the result of “↑” genes while B is the result of “↓” genes. Frequency of each term was color coded ranging from white for scarce terms to deep colors saturating with increasing frequency. Rows indicated the enriched terms and columns the frequency of the respective clusters. There was no significant enrichment found by comparisons of CF4abvs CF4ac and CF18acvs control, thus these two comparisons were not included in the figure.