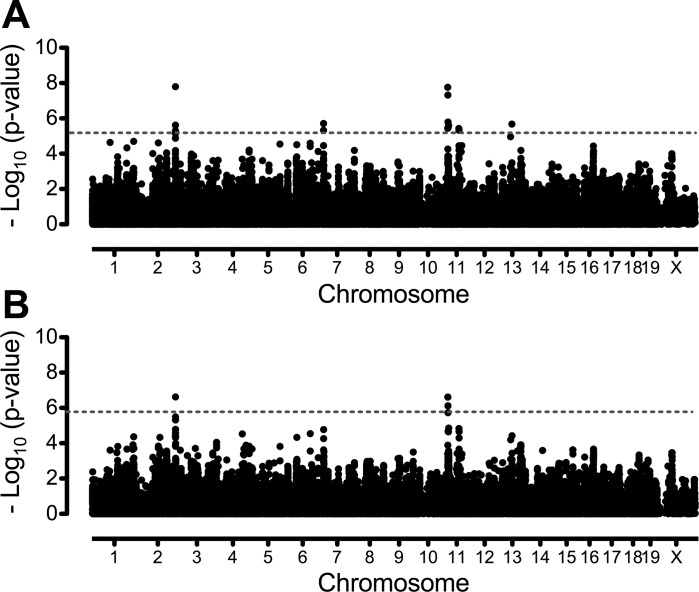
Fig. 3.
Genome-wide association mapping for endurance exercise capacity, expressed as time in minutes, in male mice from 34 classical and wild-derived inbred strains (A) and 30 classical inbred strains only (B) using the 132 K SNP panel. Association mapping was conducted with an empirical mixed model algorithm (EMMA) and a 132,285 SNP panel. The x-axis indicates genomic position divided by chromosome. Values on the y-axis are P values transformed using −log10 (P value). Horizontal dashed lines in each plot indicate genome-wide significance thresholds. Significance thresholds were calculated using a 5% false discovery rate and correspond to P values of 7.21 × 10−6 and 1.87 × 10−6 for 34 strain and 30 strain cohorts, respectively.