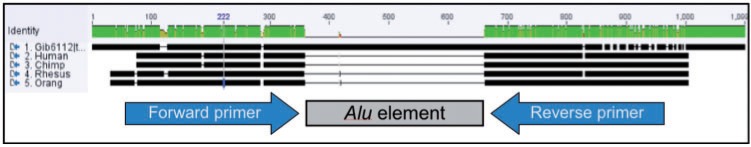
Fig. 2.
An illustration of how multiple alignments of orthologous loci are used to design oligonucleotide primers for lineage-specific insertions. The multiple alignment was performed using Clustal X. The gibbon sequence is N. leucogenys and contains a gibbon-specific Alu insertion (indicated by the 300 bp region in the center that does not match the other sequences). Also included in this alignment are H. sapiens, P. troglodytes, Pongo abelii, and Macaca mulatta. The gray rectangle shows the location of the Alu insertion in the gibbon. The arrows indicate the orthologous flanking regions in which primers are designed that will match all five species in the alignment.