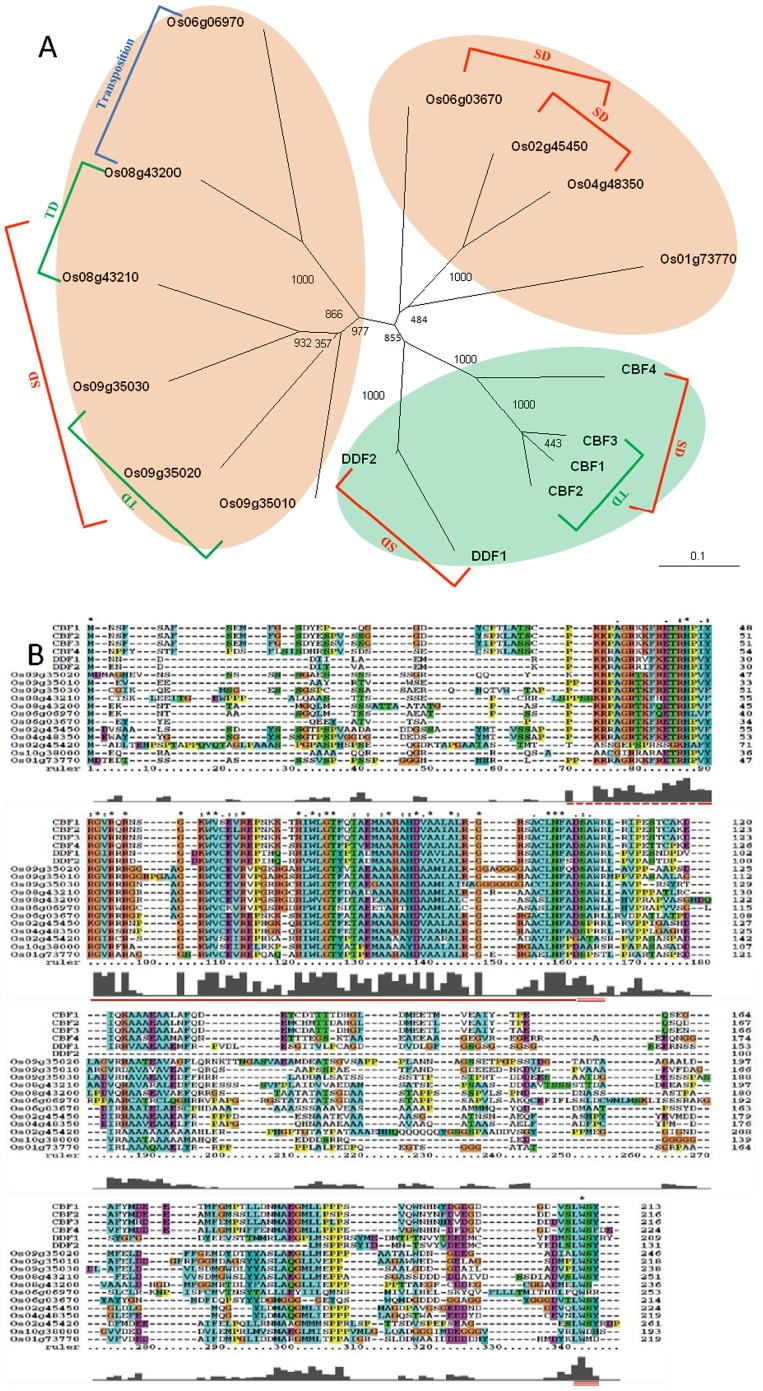
Figure 2. Phylogenetic analysis and sequence alignment of Arabidopsis DREB1s and their homologs in rice using ClustalW2.
A: The phylogenetic analysis was carried out by the neighbour-joining method of ClustalW2, and the tree was edited and viewed by TreeView software. SD: segmental duplication; TD: tandem duplication; Transposition: single gene duplication by transposition. Bootstrap values from 1000 replicates were indicated at each node. Scale bar represented 0.1 amino acid substitution per site. B: The NLS (nuclear localization signal), ERF/AP2 domain, DSAW motif, and LWSY motif are shown. The alignment of the 141–171 region of CBF1 and the corresponding regions of its rice and Arabidopsis homologs has been omitted due to the absence of conserved motif.