Figure 3.
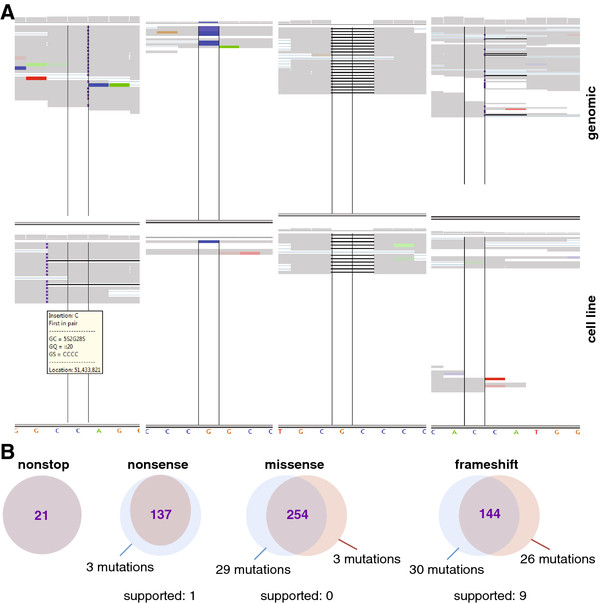
Most variant calls within genes are shared between genomic DNA and cell line. A: The Integrated Genomics Viewer (IGV) was used to assess variants in genes that were affected by non-synonymous mutations in both genomes, but where the number or the position of the variants differed. The following scenarios were encountered: (i) one of the calls in the two genomes was wrongly reported due to ambiguity in the alignments (left panel), (ii) coverage in one of the two genomes was very low at the called position, thus no variant call could be made (second left panel), (iii) a variant was determined homozygous in one of the genomes and heterozygous in the other, even though both genomes were homozygous (second right panel, “false positives”) or (iv) the two genomes looked really different (right panel, “true positive”). All reads were displayed in IGV. Each horizontal strip represents one read. Bases in agreement with the reference genome are displayed in grey, non-according bases are colored. Insertions are depicted by a purple square, deletions by a thick line, and gaps by a thin line. At the top of each panel, the relative coverage of each base in indicated by the height of the grey bar. The variant position is framed by two vertical lines. The genomic DNA is shown in the upper part of the panel, the cell line underneath it, at the bottom the reference sequence is displayed. B: For each class of non-synonymous variants (nonstop, nonsense, missense, frameshift), most mutations of this class in both genomic (blue) and cell line DNA (red) are shared between the two genomes (purple). A high percentage of the mutations called to be unique to either of the two genomes is not supported by actual reads.